The Guizetti lab over the years
Dr. Julien Guizetti (Group leader, 2017 - 2024)
Sultan Bekbayev (MSc rotation, 2023 - 2024)
Anja Klemmer (PhD student, 2019 - 2024)
Yannik Voß (MSc thesis, 2018; PhD student, 2019 - 2023)
Nicolas Lichti (BSc thesis, 2019; MSc thesis, 2022 - 2023)
Christoph Wenz (MSc rotation, 2021; MSc thesis, 2022 - 2023)
Vanessa Stürmer (intern, 2018-2019; MSc rotation, 2020 - 2021; MSc thesis, 2022 - 2023)
Sophie Stopper (MSc rotation, 2022)
Caroline Simon (PhD student, 2018 - 2022)
Marius Flörchinger (BSc thesis, 2021; MSc rotation, 2021)
Johanna Bauer (BSc thesis, 2020; MSc rotation, 2021)
Dr. Tatiany Patricia Romão (Visiting Scientist, 2020 - 2021)
Nathan Ribot (MSc rotation, 2020)
Ann-Kathrin Mehnert (MSc rotation, 2018, MSc thesis, 2019)
Marlen Wesemeyer (BSc thesis, 2018)
Nadja Ilkenhans (BSc thesis, 2018)
& various BSc students

Research focus of the Guizetti lab while in Heidelberg
The severity of the malaria disease, which still causes the death of about 600’000 people per year, is predicted by the number of Plasmodium-infected red blood cells that circulate in the blood stream. Yet, we only have a rudimentary understanding about the unconventional cell division mechanism the parasite uses for its rapid proliferation. Contrary to most model organisms, which divide by binary fission, malaria-causing parasites first multiply their nuclei and later generate 8-30 daughter cells at once (Fig. 1). I
n the Guizetti group we study the mechanism of nuclear division to ultimately uncover how the parasite achieves its final number of daughter cells. We use modern super-resolution, electron, and live cell microscopy technologies combined with CRISPR/Cas9 genome editing to describe the kinetics of nuclear multiplication, study the organization and composition of the centrosome, and aim to uncover the putative role of phase separation in centrosome function (see projects). By studying this divergent eukaryote, we are contributing an evolutionary cell biology perspective, but also aim to uncover new targets within this essential pathway that can be leveraged in the fight against malaria.
-

In "simple" words:
Malaria is a serious disease that still kills more than half a million people every year. The disease develops when the malaria pathogen, called Plasmodium falciparum, multiplies massively in the blood of an infected person. After an infectious mosquito bite and a stopover in the liver, the small pathogen penetrates our red blood cells. There it digests the blood cell from the inside and grows. In the second half of this development cycle, it begins to multiply its own cell nuclei, which contain the genetic material in the form of DNA. Only at the end of its development does it pack the cell nuclei into 8 to 30 daughter cells, which then escape from the bursting host cell to infect many new red blood cells. This multiplication process differs significantly from that which takes place in our body cells, for example, which usually undergo a two-part division. This is another reason why we understand the cell division of the parasite very little. The aim of the Guizetti research group is to use modern microscopy techniques to investigate and understand this process with a high temporal and spatial resolution (see films and images). We hope to uncover new target molecules for intervention strategies in the fight against malaria.
More information on malaria can be found in our service area!
Projects
Cell division is a highly dynamic process. In the case of malaria parasites, key cellular parameters have, however, not been assessed in a time-resolved manner. We develop time-lapse imaging approaches combined with live cell staining protocols to determine division timing, nuclear multiplication rates, and spindle reorganization and more at the level of individual parasites (Fig. 2). We are thereby creating a quantitative cell biological framework that will enable the study of malaria parasite cell division with unprecedented spatial and temporal resolution.
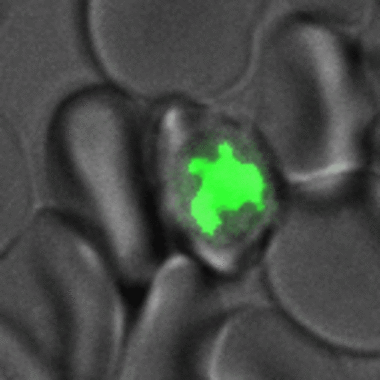
Figure 2. Malaria-parasites produce multiple nuclei before egressing from host cell. Time lapse movie of parasite infected red blood cell from start of nuclear division until bursting and reinvasion of neighboring cell. Red blood cell and parasite can be seen in transmission channel (grey), while the chromatin, i.e. the nuclei, of the parasite are fluorescently labeled by expression of GFP-tagged Histone 2B (green).
Nuclear division requires small and intricate molecular machines that physically pull apart chromosomes after their replication. Key components of this machinery are i) the mitotic spindle, which consists of tube-shaped polymers called microtubules, to which individual chromosomes are attached and ii) the centrosomes which organizes the microtubules and form the two poles to which the chromosomes are pulled (Fig. 3). The organization of the surprisingly small mitotic spindle and the atypical centrosome in Plasmodium has not been well studied. Using a combination of super-resolution and electron microscopy we have created the first detailed working model of the malaria parasite centrosome (Fig. 3). We can now use this model to step-by-step identify the components of this divergent organelle and elucidate their function.

Figure 3. Working model of the malaria parasite centrosome and mitotic spindle. Microtubules (magenta) form a bipolar array called mitotic spindle. The spindle pole, also called centrosome (grey), is a protein dense structure that spans the nuclear membrane (grey blue) through a special nuclear pore (red). The extranuclear part contains centrins (green) one of the few conserved centrosomal protein families. The intranuclear part harbors microtubule nucleation site (white) from which microtubules extend towards the nuclear DNA (blue). Later this mitotic spindle will extend to drive apart the sister chromosomes. Adapted from Simon et al. 2021 (PMID: 34535568).
Centrosomes can be viewed as membrane-less organelles that concentrate many different components in a defined sub-cellular compartment. In recent years the new biophysical concept of phase separation, which describes the propensity of certain proteins to coacervate under specific conditions, has been employed to explain the properties of membrane-less organelles including the centrosomes. Phase-separation has not yet been investigated in malaria-causing parasites, but when studying recombinant P. falciparum centrins, which are conserved calcium-binding proteins, in vitro we found that some can undergo liquid-liquid phase separation (Fig. 4). The importance of this feature for centrosome assembly and function in vivo is the subject of further studies in the group.
Figure 4. Recombinant centrin protein droplets display liquid-like behavior. Transmission time-lapse microscopy of a recombinant PfCen1 protein solution after addition of calcium. Fusion between two protein droplets indicates that they constitute liquid-liquid phase condensates rather than solid protein assemblies. Adapted from Voss et al. 2022 (biorxiv.org/content/10.1101/2022.07.26.501452v1).
Movie gallery
Movie 1 - Malaria parasites generates multiple daughter cells that invade neighboring erythrocytes.
Movie 2 - Highly resolved three-dimensional organization of microtubules and centrosomes in multinucleated malaria parasite.
Further movies
Movie 3 - Multiple rounds mitotic spindle formation and extension malaria parasite.
Movie 5 - Three-dimensional organization of microtubules, nuclear pores, and mitotic spindle in dividing nuclei using ultrastructure expansion microscopy.
Movie 4 - Three-dimensional organization of microtubules, centriolar plaques, nuclear pores and hemispindle in dividing nuclei using ultrastructure expansion microscopy.
Publications / Collaborations
Original articles
Voß Y, Klaus S, Lichti NP, Ganter M, Guizetti J (2023) Malaria parasite centrins can assemble by Ca2+-inducible condensation. PLoS Pathog. Dec 27;19(12):e1011899. doi: 10.1371/journal.ppat.1011899. PMID: 38150475; PMCID: PMC10775985.
Machado M, Klaus S, Klaschka D, Guizetti J, Ganter M (2023) Plasmodium falciparum CRK4 links early mitotic events to the onset of S-phase during schizogony. mBio. Jun 22:e0077923. doi: 10.1128/mbio.00779-23. Epub ahead of print. PMID: 37345936.
Wenz C, Simon CS, Romão TP, Stürmer VS, Machado M, Klages N, Klemmer A, Voß Y, Ganter M, Brochet M, Guizetti J (2023) An Sfi1-like centrin-interacting centriolar plaque protein affects nuclear microtubule homeostasis. PLoS Pathog. May 2;19(5):e1011325. doi: 10.1371/journal.ppat.1011325. PMID: 37130129; PMCID: PMC10180636.
Ganter M*, Guizetti J*, Kilian N* (2022) Visualization of Infected Red Blood Cell Surface Antigens by Fluorescence Microscopy In: Jensen ATR, Hviid L, editors. Malaria Immunology: Targeting the Surface of Infected Erythrocytes. New York, NY: Springer US. pp. 425–433. doi:10.1007/978-1-0716-2189-9_31
Voss Y, Klaus S, Ganter M, Guizetti J* (2022) Ca2+-inducible phase separation of centrins in proliferating malaria parasites. bioRxiv 2022.07.26.501452. doi:10.1101/2022.07.26.501452
Klaus S, Binder P, Kim J, Machado M, Funaya C, Schaaf V, Klaschka D, Kudulyte A, Cyrklaff M, Laketa V, Höfer T, Guizetti J, Becker NB, Frischknecht F, Schwarz US, Ganter M (2022) Asynchronous nuclear cycles in multinucleated Plasmodium falciparum facilitate rapid proliferation. Sci Adv. Apr;8(13):eabj5362. doi: 10.1126/sciadv.abj5362. Epub 2022 Mar 30. PMID: 35353560.
Machado M, Klaus S, Klaschka D, Guizetti J, Ganter M (2022) Plasmodium falciparum CRK4 links early mitotic events to the onset of S-phase during schizogony. bioRxiv 2022.08.31.505163. doi:10.1101/2022.08.31.505163.
Wenz C, Simon CS, Romão TP, Stürmer V, Machado M, Klages N, Klemmer A, Voß Y, Ganter M, Brochet M, Guizetti J* (2022) An Sfi1-like centrin-interacting centriolar plaque protein affects nuclear microtubule homeostasis. bioRxiv 2022.07.28.501831. doi:10.1101/2022.07.28.501831
Diehl M, Roling L, Rohland L, Weber S, Cyrklaff M, Sanchez CP, Beretta CA, Simon CS, Guizetti J, Hahn J, Schulz N, Mayer MP, Przyborski JM (2021) Co-chaperone involvement in knob biogenesis implicates host-derived chaperones in malaria virulence. PLoS Pathog. Oct 6;17(10):e1009969. doi: 10.1371/journal.ppat.1009969. PMID: 34614006; PMCID: PMC8544838.
Simon CS, Funaya C, Bauer J, Voβ Y, Machado M, Penning A, Klaschka D, Cyrklaff M, Kim J, Ganter M, Guizetti J (2021) An extended DNA-free intranuclear compartment organizes centrosome microtubules in malaria parasites. Life Sci Alliance. Sep 17;4(11):e202101199. doi: 10.26508/lsa.202101199. PMID: 34535568; PMCID: PMC8473725.
Simon CS, Stürmer VS, Guizetti J (2021) How Many Is Enough? - Challenges of Multinucleated Cell Division in Malaria Parasites. Front Cell Infect Microbiol. May 7;11:658616. doi: 10.3389/fcimb.2021.658616. PMID: 34026661; PMCID: PMC8137892.
Soni K, Kempf G, Manalastas-Cantos K, Hendricks A, Flemming D, Guizetti J, Simon B, Frischknecht F, Svergun DI, Wild K, Sinning I (2021) Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation. Commun Biol. May 20;4(1):600. doi: 10.1038/s42003-021-02132-y. PMID: 34017052; PMCID: PMC8137916.
Guizetti J, Frischknecht F (2021) Apicomplexans: A conoid ring unites them all. PLoS Biol. Mar 11;19(3):e3001105. doi: 10.1371/journal.pbio.3001105. PMID: 33705378; PMCID: PMC7951970.
Barcons-Simon A, Cordon-Obras C, Guizetti J, Bryant JM, Scherf A. (2020) CRISPR Interference of a Clonally Variant GC-Rich Noncoding RNA Family Leads to General Repression of var Genes in Plasmodium falciparum. mBio. Jan 21;11(1)
Mehnert, A.K., Simon C.S., Guizetti, J. (2019). Immunofluorescence staining protocol for STED nanoscopy of Plasmodium-infected red blood cells. Mol Biochem Parasitol. 229, 47–52.
Müller, L. S. M., Cosentino, R. O., Förstner, K. U., Guizetti, J., Wedel, C., Kaplan, N., … Siegel, T. N. (2018). Genome organization and DNA accessibility control antigenic variation in trypanosomes. Nature 563(7729), 121–125
Zanghì, G., Vembar, S. S., Baumgarten, S., Ding, S., Guizetti, J., Bryant, J. M., … Scherf, A. (2018). A Specific PfEMP1 Is Expressed in P. falciparum Sporozoites and Plays a Role in Hepatocyte Infection. Cell Reports 22(11), 2809–2817.
Bryant J.M., Regnault C., Scheidig-Benatar C., Baumgarten S., Guizetti J.*, Scherf A. (2017). CRISPR/Cas9 Genome Editing Reveals That the Intron Is Not Essential for var2csa Gene Activation or Silencing in Plasmodium falciparum. MBio 8(4). pii: e00729-17.
Guizetti, J.*, Barcons-Simon, A., Scherf, A. (2016). Trans-acting GC-rich non-coding RNA at var expression site modulates gene counting in malaria parasite. Nucleic Acids Res 44, 9710–9718.
Zhang, Q., Siegel, T. N., Martins, R. M., Wang, F., Cao, J., Gao, Q., Cheng, X., Jiang, L., Hon, C. C., Scheidig-Benatar, C., Sakamoto, H., Turner, L., Jensen, A. T., Claes, A., Guizetti, J., Malmquist, N. A., and Scherf, A. (2014). Exonuclease-mediated degradation of nascent RNA silences genes linked to severe malaria. Nature 513, 431–435.
Guizetti, J., Martins, R. M., Guadagnini, S., Claes, A., and Scherf, A. (2013). Nuclear Pores and Perinuclear Expression Sites of var and Ribosomal DNA Genes Correspond to Physically Distinct Regions in Plasmodium falciparum. Eukaryot Cell 12, 697–702.
Guizetti J*, Scherf A. (2013). Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell Microbiol. doi:10.1111/cmi.12115.
Guizetti J, Gerlich DW. (2012). ESCRT-III polymers in membrane neck constriction. Trends Cell Biol 22:133–140. doi:10.1016/j.tcb.2011.11.007.
Guizetti, J., Schermelleh, L., Mantler, J., Maar, S., Poser, I., Leonhardt, H., Muller-Reichert, T., and Gerlich, D. W. (2011). Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331, 1616–1620.
Guizetti, J., Mantler, J., Muller-Reichert, T., and Gerlich, D. W. (2010). Correlative time-lapse imaging and electron microscopy to study abscission in HeLa cells. Methods Cell Biol 96, 591–601.
Lacroix, B., van Dijk, J., Gold, N. D., Guizetti, J., Aldrian-Herrada, G., Rogowski, K., Gerlich, D. W., and Janke, C. (2010). Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J Cell Biol 189, 945–954.
Guizetti J, Gerlich DW. (2010). Cytokinetic abscission in animal cells. Semin Cell Dev Biol 21:909–916. doi:10.1016/j.semcdb.2010.08.001.
Guizetti J, Mäntler J, Müller-Reichert T, Gerlich DW. (2010). Correlative time-lapse imaging and electron microscopy to study abscission in hela cells. Methods Cell Biol 96:591–601. doi:10.1016/S0091-679X(10)96024-X.
Lekomtsev, S., Guizetti, J., Pozniakovsky, A., Gerlich, D. W., and Petronczki, M. (2010). Evidence that the tumor-suppressor protein BRCA2 does not regulate cytokinesis in human cells. J Cell Sci 123, 1395–1400.
Steigemann, P., Wurzenberger, C., Schmitz, M. H., Held, M., Guizetti, J., Maar, S., and Gerlich, D. W. (2009). Aurora B‑mediated abscission checkpoint protects against tetraploidization. Cell 136, 473–484.
Reviews
Voß Y, Klaus S, Guizetti J, Ganter M. (2023) Plasmodium schizogony, a chronology of the parasite's cell cycle in the blood stage. PLoS Pathog. Mar 2;19(3):e1011157. doi: 10.1371/journal.ppat.1011157. PMID: 36862652; PMCID: PMC9980825.
Guizetti, J.*, and Scherf, A. (2013). Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell Microbiol 15, 718–726.
Guizetti, J., and Gerlich, D. W. (2012). ESCRT-III polymers in membrane neck constriction. Trends Cell Biol 22, 133–140.
Guizetti, J., and Gerlich, D. W. (2010). Cytokinetic abscission in animal cells. Semin Cell Dev Biol 21, 909–916.
External collaborators
Mathieu Brochet, University of Geneva
Paul Guichard and Virginie Hamel, University of Geneva
Robert Moon, London School of Hygiene and Tropical Medicine
Moritz Treeck, Francis Crick Institute London
Kai Johnsson, MPI for Medical Research Heidelberg
Sebastian Baumgarten and Jessica Bryant, Institut Pasteur Paris
Group activities
News - Guizetti lab
New paper in "Life Science Alliance"
(publication date: 17 September 2021)
In the new article the Guizetti lab and colleagues provide a first model of the atypical parasite centrosome, which is essential for orchestrating the rapid cell division of the Plasmodium parasite causing malaria:
"Moment for Research"
(Online lecture by our group leader Julien)
As part of the Daimler and Benz Foundation's monthly seminar series "Moment for Research", our group leader Julien reports on his research in a 15-minute online lecture entitled "Malaria Pathogens in the Blood – Multiplying and Hiding" (Please note: the lecture is in German). The series offers insights into current research topics funded by the Daimler and Benz Foundation. The protagonists are scholarship holders of the postdoctoral funding programme who present their individual projects. So there is something exciting to discover every month!