Research Group Müller
Research projects in our group focus on "Dynamic Events in HIV‑1 Replication".
Projects
We are interested in the biology of the human immunodeficiency virus (HIV‑1). With our work, we want to contribute to a detailed understanding of the intricate interactions of this important pathogen with its host cell during the viral replication cycle.
The interaction of a virus with its host cell is a highly dynamic process, involving ordered and regulated formation, transport, transformation and dissociation of (nucleo)protein complexes. Biochemical, electron microscopic and structural studies provide detailed images of HI virions and information about the composition of functionally important subviral complexes. However, these methods yield snapshots or ensemble data, which do not reflect the dynamics of individual events occurring in the infected cell. In contrast, modern fluorescence imaging techniques enable us to directly observe transport processes and protein interactions within living cells. Furthermore, super-resolution and correlative microscopy now bridge the gap between electron and light microscopy, allowing the visualization of subviral details via fluorescence microscopy. All these advanced imaging methods require attachment of fluorescent labels to the viral protein of interest.
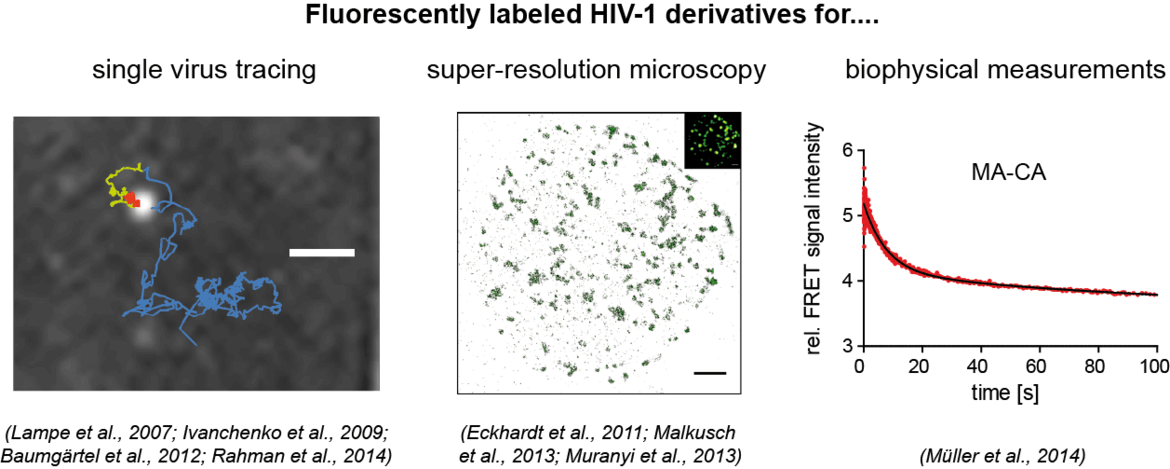
Our aim is to analyze dynamic processes in HIV‑1 replication in a quantitative manner. For this, we develop fluorescently labeled HIV‑1 derivatives and fluorescent probes, which allow us to follow individual events in virus-cell interaction with high time resolution. These studies are combined with biochemical and virological analyses. In our current work, we particularly focus on the processes involved in the formation of the infectious HIV‑1 capsid by proteolytic maturation, and on the fate and role of this capsid structure upon entry of the virus into a new host cell. See also: Movie on our Research Work
HIV‑1 Assembly and Maturation
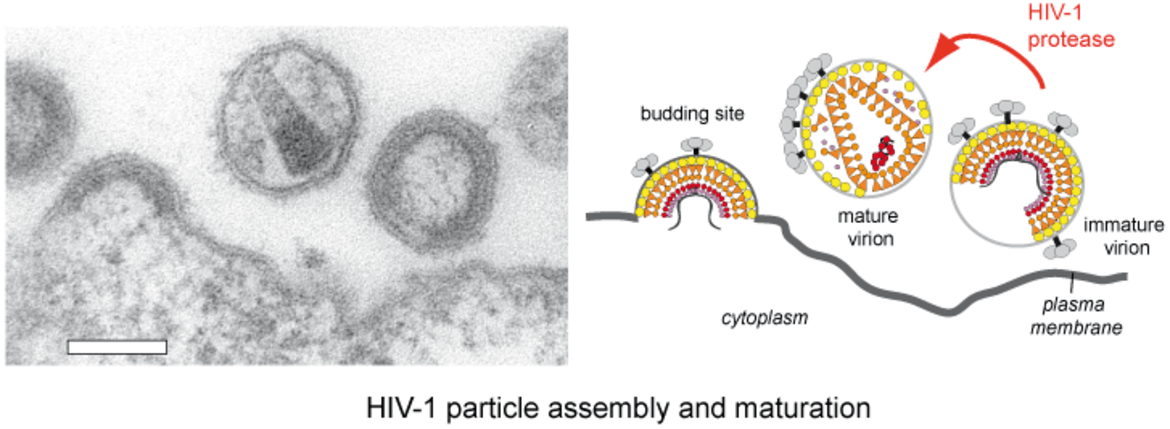
HIV‑1 particles are released from the host cell as immature non-infectious virions. They only become infectious after undergoing a complex series of proteolytic maturation steps, in which the viral polyproteins Gag and GagPol are cleaved at multiple sites in an ordered sequence by the viral protease. This proteolytic processing is accompanied by dramatic rearrangements of the virus architecture. It is well established that the process of proteolytic and structural maturation needs to be tightly regulated to achieve formation of infectious virus. Precise characterization of this regulation is crucial for our understanding of HIV‑1 morphogenesis.
Despite intense research on this topic, a number of important questions are still unanswered. This includes very basic questions as:
- when, where and how is maturation initiated?
- what are the structural intermediates?
- what is the time course of proteolytic and structural maturation?
Our aim is to develop a better understanding of the complex and dynamic events occurring during HIV‑1 particle assembly, polyprotein processing and structural maturation. For this, we develop novel fluorescence-based readout systems to monitor protease activation or activity, and combine biochemical and virological approaches with microscopic analyses.
Unexpectedly, not only the capsid structure of HIV-1, but also the matrix lattice undergoes a complex structural rearrangement during maturation, suggesting that the matrix lattice plays a role in the early viral replication steps. Together with our collaborators, we want to elucidate this enigmatic function of the HIV-1 MA protein.
Dynamics of HIV‑1 Post-Entry Events
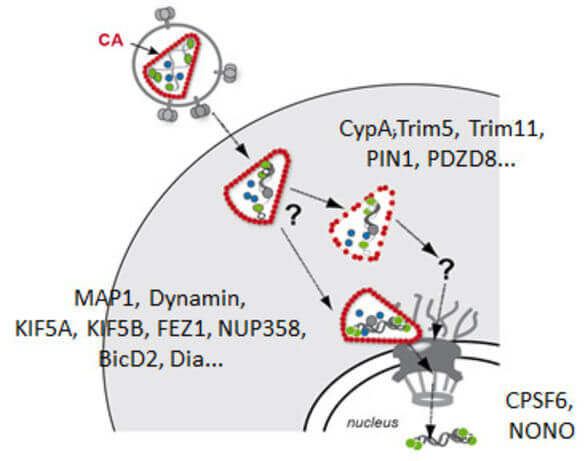
Early post-entry events, from cytoplasmic entry of the viral core to integration of the viral genome into the host DNA, represent the most enigmatic steps in the HIV‑1 replication cycle. Fusion of the viral envelope with the cell membrane releases the capsid, which encases the ssRNA genome, into the cytoplasm. The viral RNA genome is converted into dsDNA by the viral reverse transcriptase and transported through the nuclear pore into the host nucleus, where it is covalently integrated into the host genome. Reverse transcription, nuclear import and integration occur within ill-characterised nucleoprotein complexes (functionally designated as reverse transcriptase complex and pre-integration complex). Uncoating of the HIV‑1 capsid is apparently functionally linked to reverse transcription and nuclear import. Therefore, events in the post-entry phase need to be very tightly controlled in time and space. Results from many labs indicate that the viral capsid plays a central role in regulating post-entry steps. An increasing number of capsid binding host cell proteins that either promote or restrict viral replication is also implicated in these events.
While these facts are well established, the sequence and intracellular localisation of molecular events, temporal and functional correlation between subsequent steps and the roles of viral and cellular proteins involved are a matter of intense debate. The study of post-entry events is complicated by the facts that (i) the subviral complexes undergo a series of dynamic transitions, (ii) events are not synchronized and a cell may contain many viral complexes in different stages, and (iii) not all entry events are productive and some – or many – lead into a dead end. Furthermore, alternative pathways appear to exist for distinct steps; these may differ between different cell types, may be used alternatively, or even occur in parallel.
Live-cell microscopy can help to overcome these obstacles, since it allows focusing on individual subviral complexes and can quantitatively describe a dynamic sequence of events with high time resolution. We are therefore developing and applying improved replication competent labeled HIV variants and novel, minimally invasive labelling strategies, in order to visualize individual events in the early stages of the HIV‑1 replication cycle using advanced microscopic methods.
Important questions we try to address in our work are:
- What is the role of viral proteins in post entry steps?
- Where and when does capsid uncoating happen?
- What is the function of specific host cell factors?
- What is the mechanism of PIC nuclear import in different host cell types?
Pandemic response
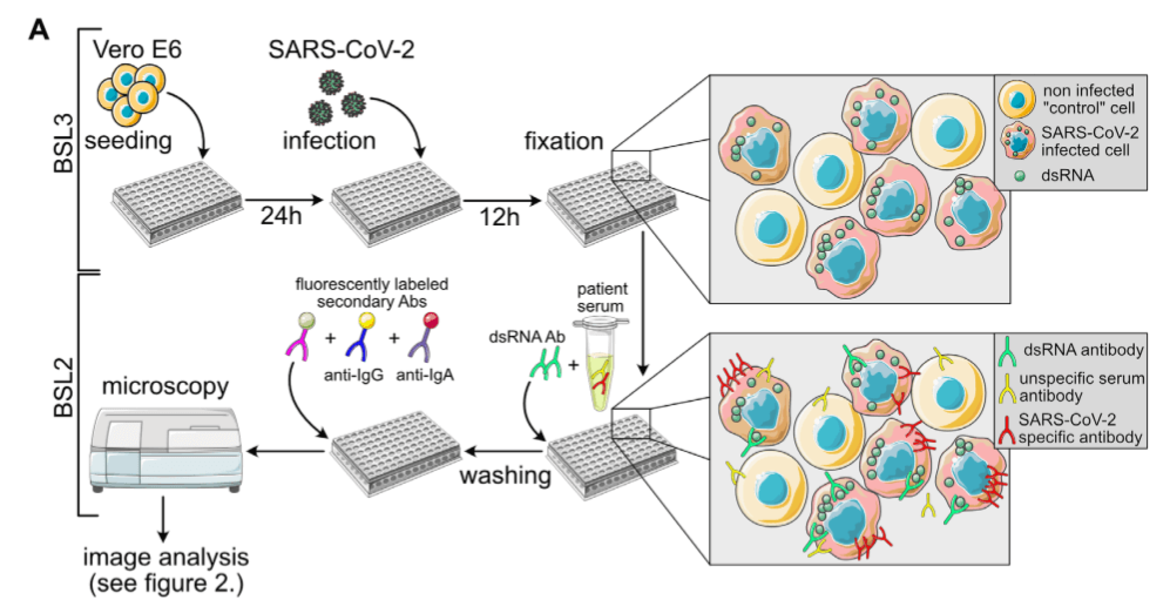
Retroviruses, in particular HIV-1, are the focus of our research work, and this will remain so. Nevertheless, we reacted to the pandemic emergency by supporting COVID-19 related clinical research projects. In the spring of 2020, we teamed up with colleagues from the CIID and bioinformaticians to develop a semi-automated, semi-quantitative immunofluorescence assay for the determination of Sars-CoV-2 specific antibodies, which was urgently needed at the time for validation experiments in epidemiological studies under conditions of low seroprevalence. We were also involved in establishing a multiplex assay for antibody measurements.
Using these and other assays our group has contributed to clinical studies, in particular the Baden-Württemberg childrens study, household study and follow-up analyses to investigate prevalence and transmission of Sars-CoV-2 within families in the first wave of the pandemic. We are currently involved in local and multi-centric studies addressing post-acute consequences of COVID-19 (long COVID) as part of the EPILOC consortium.
Selected Publications
Complete Publication List (PubMed)
ORCID 0000-0001-5726-5585
*** Co-Corresponding Authors
- Peter RS Nieters A Göpel S Merle U Steinacker JM Deibert P Friedmann-Bette B Nieß A Müller B Schilling C Erz G Giesen R Götz V Keller K Maier P Matits L Parthé S Rehm M Schellenberg J Schempf U Zhu M Kräusslich HG Rothenbacher D Kern WV EPILOC Phase 2 Study GroupPeter RS Nieters A Göpel S Merle U Steinacker JM Deibert P Friedmann-Bette B Nieß A Müller B Schilling C Erz G Giesen R Götz V Keller K Maier P Matits L Parthé S Rehm M Schellenberg J Schempf U Zhu M Kräusslich HG Rothenbacher D Kern WV EPILOC Phase 2 Study Group. (2025) Persistent symptoms and clinical findings in adults with post-acute sequelae of COVID-19/post-COVID-19 syndrome in the second year after acute infection: A population-based, nested case-control study. PLoS Medicine. 22(1): e1004511. doi: 10.1371/journal.pmed.1004511
- Sartingen N, Stürmer V, Kaltenböck M, Müller TG, Schnitzler P, Kreshuk A, Kräusslich HG, Merle U, Mücksch F, Müller B, Pape C, Laketa V. (2024) Multiplex Microscopy Assay for Assessment of Therapeutic and Serum Antibodies against Emerging Pathogens. Viruses , 16(9), 1473; https://doi.org/10.3390/v16091473
- Ananth S, Ambiel I, Schifferdecker S, Müller TG, Wratil PR, Mejias-Perez E, Kräusslich HG, Müller B, Keppler OT, Fackler OT. (2024). Spatial resolution of HIV-1 post-entry steps in resting CD4 T cells. Cell Rep. 2024 Mar 12;43(3):113941. doi: 10.1016/j.celrep.2024.113941
- Schifferdecker S, Zila V, Müller TG, Sakin V, Anders-Össwein M, Laketa V, Kräusslich HG, Müller B (2022). Direct Capsid Labeling of Infectious HIV-1 by Genetic Code Expansion Allows Detection of Largely Complete Nuclear Capsids and Suggests Nuclear Entry of HIV-1 Complexes via Common Routes. mBio. doi: 10.1128/mbio.01959-22
- Volpato A, Ollech D, Alvelid J, Damenti M, Müller B, York AG, Ingaramo M, Testa I. (2022). Extending fluorescence anisotropy to large complexes using reversibly switchable proteins. Nat Biotechnol. doi: 10.1038/s41587-022-01489-7
- Müller, TG, Zila V, Müller B, Kräusslich HG (2022) Nuclear Capsid Uncoating and Reverse Transcription of HIV-1. Annual Reviews of Virology 9:20.1–20.24. https://doi.org/10.1146/annurev-virology-020922-110929
- Qian C., Flemming A., Müller B.***, Lamb DC***. (2022) Dynamics of HIV-1 Gag Processing as Revealed by Fluorescence Lifetime Imaging Microscopy and Single Virus Tracking. Viruses 14, 340. doi.org/10.3390/v14020340
- Renk, H., Dulovic, A., Seidel, A., Becker, M., Fabricius, D., Zernickel, M., Junker, D., Gross, R., Muller, J., Hilger, A., et al. (2022). Robust and durable serological response following pediatric SARS-CoV-2 infection. Nature communications 13, 128.
- Zila V, Müller TG, Müller B, Kräusslich HG (2021) HIV-1 capsid is the key orchestrator of early viral replication. PLoS Pathog 17(12):e1010109. doi: 10.1371/journal.ppat.1010109.
- Qu, K., Ke, Z., Zila, V. Anders-Össwein, M., Glass, B., Mücksch, F., Müller, R., Schultz, C., Müller, B., Kräusslich, H.G. and Briggs, J.A.G. (2021) Maturation of the matrix and viral membrane of HIV-1. Science, Vol. 373, Issue 6555, pp. 700-704. DOI: 10.1126/science.abe6821
- Müller TG, Zila V, Peters K, Schifferdecker S, Stanic M, Lucic B, Laketa V, Lusic M, Müller B, Kräusslich HG. (2021) HIV-1 uncoating by release of viral cDNA from capsid-like structures in the nucleus of infected cells. Elife. 10:e64776. doi: 10.7554/eLife.64776. Epub ahead of print. PMID: 33904396.
- Seeßle, J., Waterboer, T., Hippchen, T., Simon, J., Kirchner, M., Lim, A., Müller, B., and Merle, U. (2021) Persistent symptoms in adult patients one year after COVID-19: a prospective cohort study. Clinical Infectious Diseases, ciab611, https://doi.org/10.1093/cid/ciab611
- Butt J, Murugan R, Hippchen T, Olberg S, van Straaten M, Wardemann H, Stebbins E, Kräusslich HG, Bartenschlager R, Brenner H, Laketa V, Schöttker B, Müller B, Merle U, Waterboer T. (2021) From Multiplex Serology to Serolomics-A Novel Approach to the Antibody Response against the SARS-CoV-2 Proteome.Viruses 13(5):749. doi: 10.3390/v13050749.
- Zila V, Margiotta E, Turoňová B, Müller TG, Zimmerli CE, Mattei S, Allegretti M, Börner K, Rada J, Müller B, Lusic M, Kräusslich HG, Beck M. (2021). Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell. S0092-8674(21)00068-4. doi: 10.1016/j.cell.2021.01.025
- Stich M, Elling R, Renk H, Janda A, Garbade SF, Müller B, Kräusslich HG, Fabricius D, Zernickel M, Meissner P, Huzly D, Grulich-Henn J, Haddad A, Görne T, Spielberger B, Fritsch L, Nieters A, Hengel H, Dietz AN, Stamminger T, Ganzenmueller T, Ruetalo N, Peter A, Remppis J, Iftner T, Jeltsch K, Waterboer T, Franz AR, Hoffmann GF, Engel C, Debatin KM, Tönshoff B, Henneke P. (2021) Transmission of SarsCoV-2 in households with children, Southwest Germany, May-August 2020. Emerg Infect Dis 27(12):3009-3019. doi: 10.3201/eid2712.210978.
- Tönshoff B, Müller B, Elling R, Renk H, Meissner P, Hengel H, Garbade SF, Kieser M, Jeltsch K, Grulich-Henn J, Euler J, Stich M, Chobanyan-Jürgens K, Zernickel M, Janda A, Wölfle L, Stamminger T, Iftner T, Ganzenmueller T, Schmitt C, Görne T, Laketa V, Olberg S, Plaszczyca A, Cortese M, Bartenschlager R, Pape C, Remme R, Huzly D, Panning M, Weigang S, Giese S, Ciminski K, Ankerhold J, Kochs G, Schwemmle M, Handgretinger R, Niemeyer CM, Engel C, Kern WV, Hoffmann GF, Franz AR, Henneke P, Debatin KM, Kräusslich HG. (2021) Prevalence of SARS-CoV-2 Infection in Children and Their Parents in Southwest Germany.JAMA Pediatr. 2021 Jan 22. doi: 10.1001/jamapediatrics.2021.0001.
- Pape, C., Remme, R., Wolny, A., Olberg, S., Wolf, S., Cerrone, L., Cortese, M., Klaus, S., Lucic, B., Ullrich, S., Anders-Össwein, A., Wolf, S., Cerikan, B., Neufeldt, C.J., Ganter, M., Schnitzler, P., Merle, U. Lusic, M., Boulant, S., Stanifer, M., Bartenschlager, R., Hampecht, F.A., Kreshuk, A., Tischer, C., Kräusslich, H.G., Müller, B.*** and Laketa, V.*** (2020). Microscopy-based assay for semi-quantitative detection of SARS-CoV-2 specific antibodies in human sera: A semi-quantitative, high throughput, microscopy-based assay expands existing approaches to measure SARS-CoV-2 specific antibody levels in human sera. Bioessays, e2000257.
- Spilger, R., Imle, A., Lee, J.Y., Müller, B., Fackler, O.T., Bartenschlager, R., and Rohr, K. (2020). A Recurrent Neural Network for Particle Tracking in Microscopy Images Using Future Information, Track Hypotheses, and Multiple Detections. IEEE Trans Image Process
- Kalusche, S., Vanshylla, K., Kleipass, F., Gruell, H., Müller, B., Zeng, Z., Koch, K., Stein, S., Marcotte, H., Klein, F., and Dietrich, U. (2020). Lactobacilli Expressing Broadly Neutralizing Nanobodies against HIV-1 as Potential Vectors for HIV-1 Prophylaxis? Vaccines (Basel) 8.
- Müller, T.G., Sakin, V., and Müller, B. (2019). A Spotlight on Viruses - Application of Click Chemistry to Visualize Virus-Cell Interactions. Molecules 24.
- Bejarano, D.A., Peng, K., Laketa, V., Börner, K., Jost, K.L., Lucic, B., Glass, B., Lusic, M., Müller, B., and Kräusslich, H.G. (2019). HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the Nuclear Pore Complex. eLife, e41800
- Imle, A., Kumberger, P., Schnellbacher, N.D., Fehr, J., Carrillo-Bustamante, P., Ales, J., Schmidt, P., Ritter, C., Godinez, W.J., Müller, B., Rohr, K., Hamprecht, F.A., Schwarz, U.S., Graw, F., and Fackler, O.T. (2019). Experimental and computational analyses reveal that environmental restrictions shape HIV-1 spread in 3D cultures. Nature communications 10, 2144.
- Mattei S., Tan A, Glass B, Müller B., Kräusslich H.G., and Briggs J.A.G. (2018) High-resolution structures of HIV-1 Gag cleavage mutants determine structural switch for virus maturation. Proc Natl Acad Sci USA.
- Sakin V, Hanne J, Dunder J, Anders-Össwein M, Laketa V, Nikić I, Kräusslich HG, Lemke EA, Müller B. (2017) A Versatile Tool for Live-Cell Imaging and Super-Resolution Nanoscopy Studies of HIV-1 Env Distribution and Mobility. Cell Chem Biol. 24: 635-645.e5.
- Mücksch F, Laketa V, Müller B, Schultz C, Kräusslich HG (2017) Synchronized HIV assembly by tunable PIP2 changes reveals PIP2 requirement for stable Gag anchoring. Elife. pii: e25287. doi: 10.7554/eLife.25287
- Hanne J, Göttfert F, Schimer J, Anders-Össwein M, Konvalinka J, Engelhardt J, Müller B, Hell SW, Kräusslich HG (2016). Stimulated Emission Depletion Nanoscopy Reveals Time-Course of Human Immunodeficiency Virus Proteolytic Maturation . ACS Nano, 10: 8215-22
- FEBS Letters Special Issue (2016) Integrative analysis of pathogen replication and spread. http://febs.onlinelibrary.wiley.com/hub/issue/10.1111/feb2.2016.590.issue-13/
- Konvalinka, J., Kräusslich, H.G., and Müller, B. (2015). Retroviral proteases and their roles in virion maturation. Virology 479-480:403-13
- Schimer J, Pavova M, Anders M, Pachl P, Sacha P, Cigler P, Weber J, Majer P, Rezacova P, Kräusslich HG, Müller B***, Konvlinka J***, (2015). Triggering HIV polyprotein processing inside virions by rapid photodegradation of a tight-binding photodestructable protease Inhibitor. Nature Communications 6:6461
- Peng K, Muranyi W., Glass B, Laketa V, Yant SR, Tsai L, Cihlar T, Müller B, Kräusslich, HG. (2014) Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. eLife 2014 10.7554/eLife.04114
- Mattei, S., Anders, M., Konvalinka, J., Kräusslich, H.G., Briggs, J.A.G. and Müller, B. (2014) Induced maturation of human immunodeficiency virus. J Virol. 88:13722-31
- Schur, F.K.M., Hagen, W.J.H., Rumlová, M., Ruml, T., Müller, B., Kräusslich, H.G., and Briggs, J.A.G. (2014) The structure of the immature HIV-1 capsid in intact . virus particles at 8.8 Å resolution. Nature. 2014 Nov 2. doi: 10.1038/nature13838
- Rahman, S.A., Koch, P., Weichsel, J., Godinez, W.J., Schwarz, U., Rohr, K., Lamb, D.C., Kräusslich, H.G., and Müller, B. (2014). Investigating the role of F-actin in human immunodeficiency virus assembly by live-cell microscopy. J Virol. 88:7904-14
- Müller, B., Anders, M., and Reinstein, J. (2014) In vitro analysis of HIV-1 particle dissociation: Gag proteolytic processing influences dissociation kinetics. PLoS ONE 9:e99504
- Müller B, Kräusslich HG (2014). HIV-1 Maturation. Springer Encyclopedia of AIDS DOI 10.1007/978-1-4614-9610-6_59-1
- Müller, B., and Krijnse-Locker, J. (2014). Imaging of HIV assembly and release. In: Methods in molecular biology, Springer Verlag, 1087, 167-184
- Müller B and Heilemann M (2013). Shedding new light on viruses: super-resolution microscopy for studying human immunodeficiency virus. Trends in Microbiology 21:522-33
- Könnyü B, Sadiq SK, Turányi T, Hírmondó R, Müller B, Kräusslich HG, Coveney PV, Müller V. 2013. Gag-Pol Processing during HIV-1 Virion Maturation: A Systems Biology Approach. PLoS Comput Biol. 9(6):e1003103.
- Muranyi, W., S. Malkusch, B. Müller, M. Heilemann, and H. G. Kräusslich.2013. Super-resolution Microscopy Reveals Specific Recruitment of HIV-1 Envelope Proteins to Viral Assembly Sites dependent on the Envelope C-Terminal Tail. PLoS Pathog, 9:e1003198
- de Marco A, Heuser AM, Glass B, Krausslich HG, Müller B*, Briggs JA*. 2012. The role of the SP2 domain and its proteolytic cleavage in HIV-1 structural maturation and infectivity. J Virol 86:13708
- Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, B. M, Hell S, Kräusslich HG. 2012. Maturation Dependent HIV-1 Surface Protein Redistribution Revealed by Fluorescence Nanoscopy. Science 2012, 338:524-528.
- Bozek K, Eckhardt M, Sierra S, Anders M, Kaiser R, Kräusslich HG, Müller B*, Lengauer T*. 2012. An expanded model for HIV-1 cell entry phenotype based on multi-parameter single-cell data. Retrovirology,
- Eckhardt M, Anders M, Muranyi W, Heilemann M, Krijnse-Locker J, Müller B. A SNAP-tagged derivative of HIV-1--a versatile tool to study virus-cell interactions. PLoS One. 2011;6(7):e22007. Epub 2011 Jul 22.
- Baumgärtel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Kräusslich HG, Bräuchle C, Müller B*, Lamb DC* (2011) Dynamics of HIV budding site interactions with an ESCRT component visualized in live cells. Nat Cell Biol,13: 469-474
- Jochmanns D, Anders M, Keuleers I, Smeulders L, Kraeusslich HG, Kraus G, Mueller B. (2010) Selective killing of human immunodeficiency virus infected cells by non-nucleoside reverse transcriptase inhibitor-induced activation of HIV protease. Retrovirology 7(1):89. [Epub ahead of print]
- Ivanchenko S, Godinez WJ, Lampe M, Kräusslich HG, Eils R, Rohr K, Bräuchle C, Müller B*, and DC Lamb (2009) Dynamics of HIV-1 Assembly and Release. PLoS Pathogens 5:e1000652
- Müller B*, Anders M, Akiyama H, Welsch S, Glass B, Nikovics K, Clavel F, Tervo HM, Keppler OT and H.G. Kräusslich. (2009) Human immunodeficiency virus Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. JBC 284:29692-703
- Carlson LA, Briggs JA, Glass B, Riches JD, Simon MN, Johnson MC, Müller B, Grünewald K, Kräusslich HG. 2008. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 4:592-9
- Lampe M, Briggs JA, Endress T, Glass B, Riegelsberger S, Kraeusslich HG, Lamb DC, Braeuchle C, Mueller B. (2007) Double-labelled HIV-1 particles for study of virus-cell interaction. Virology 360, 92-104. Epub 2006 Nov 9.
- Mueller B, Daecke J, Fackler OT, Dittmar MT, Zentgraf H, Kraeusslich HG (2004) Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J Virol 78, 10803-10813.
- von Schwedler UK, Stuchell M, Mueller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Kraeusslich HG, Kaplan J, Morham SG, Sundquist WI (2003) The protein network of HIV budding. Cell 114:701-13.
- Gross I, Hohenberg H, Wilk T, Wiegers K, Grattinger M, Mueller B, Fuller S, Kraeusslich HG (2000) A conformational switch controlling HIV-1 morphogenesis. EMBO J 19:103-113.
Group Members
apl. Prof. Dr. Barbara Müller, Principal Investigator
e-mail
CIID Room 305, Phone: +49 6221 56-1325
Claudia Unger-Bloor, BSc, secretary
e-mail
CIID Room 307, Phone: +49 6221 56-32563
Maria Anders-Össwein, Technician (MTA)
e-mail
CIID Room 324, Phone: +49 6221 56-1324
Kamil Lalik, PhD Student
e-mail
CIID Room 304, Phone: +49 6221 56-1324
Lisa Regner, MD Student
Nuno Sartingen, MD Student
Dr. Anja Schollmeier, postdoc
e-mail
CIID Room 304, Phone: +49 6221 56-1325
Snehith Dyavari Shetty, PhD Student
e-mail
CIID Room 304, Phone: +49 6221 56-1324
Alumni
Former Lab Members
- Jessica Dunder, Masters Student
- Manon Eckhardt, PhD Student and Postdoc
- Annica Flemming, PhD Student and Postdoc
- Anke-Mareil Heuser, Technician
- Nadja Ilkenhans, Master student
- Dr. Christa Kuhn, Guest Scientist
- Marko Lampe, PhD Student
- Liridona Maliqi, Bachelor Student
- Anja Meier, Diploma Student
- Denise Müller, Technician
- Oliver Meub, Diploma Student
- Sheikh Abdul Rahman, PhD Student
- Anna Carydis Ramirez Suarez, Guest Scientist (CIGB Havanna)
- Clarissa Reinbott, Bachelor Student
- Rene Sahm, PhD Student
- Dr. Volkan Sakin, Postdoc
- Anna-Lena Schäfer, Bachelor Student
- Sandra Schifferdecker, PhD student and postdoc
- Christina Schulte-Huxel, Bachelor Student
- Stephanie Ullrich, PhD student
- Katja Willrodt, Master Student
Group Leader
apl. Prof. Dr. rer. nat. Barbara Müller
Lehrkoodination Propädeutischer Block
This group is located on the 3rd floor of the Center for Integrative Infectious Disease Research (CIID) at INF 344.

