Research group Mücksch
Research projects in our group focus on “HIV-1 infection and latency”

Projects
The Mücksch lab is interested in gaining a better understanding of the interplay between HIV-1 and its host cells during HIV-1 infection and latency at the cellular and molecular level.
HIV infection remains one of the world’s most relevant infectious diseases with more than 38 million people worldwide living with HIV. Since the beginning of the HIV epidemic, more than 84 million people have been infected with HIV and about 40 million have died of HIV.
Intensive research has led to the development of antiretroviral therapy (ART), which has since been applied to control HIV infection in people living with HIV. However, despite available therapies, HIV infection still cannot be cured, resulting in a need for life-long ART. This imposes severe challenges on people living with HIV, such as limited access to ART, strict medication schedule, stigma, side-effects, and inflammation-associated co-morbidities.
The absence of a cure for HIV infection results from the formation of a latent reservoir which cannot be targeted by current therapies. HIV integrates into the host cell genome where it can persist as a latent provirus during antiretroviral treatment. Treatment interruption inevitably leads to viral rebound. With this, the latent reservoir presents the main challenge to the development of a cure for HIV infection and cure can thus only result from a detailed understanding of these processes.
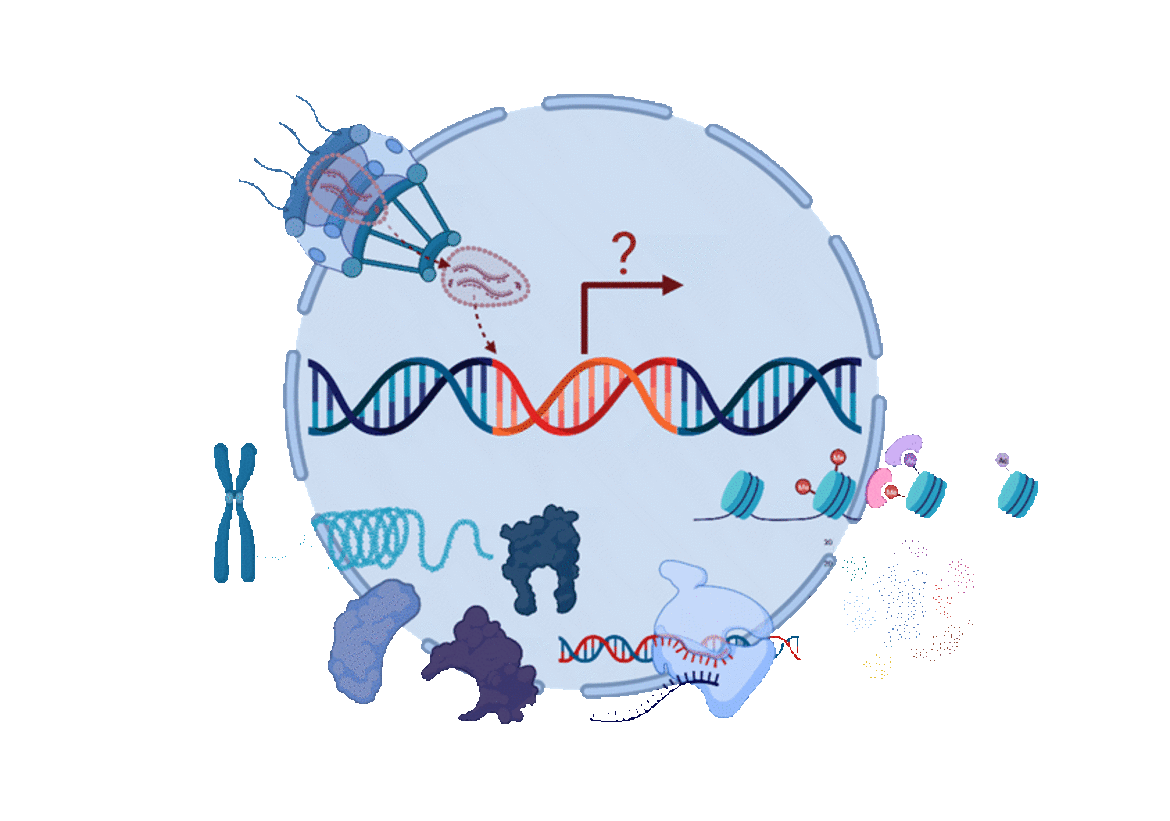
The overall aim of the Mücksch lab is to study and understand the mechanisms underlying latent HIV-1 infection. Our research goals are to characterize the heterogeneity of HIV-1 latency in cell models and primary cells, investigate how HIV enters and reverses from latency, and define the viral and host factors responsible for these processes.
We believe that a process as heterogeneous and multi-factorial as HIV-1 latency can only be studied using diverse tools and models. We are applying a proviral latency reporter, which allows for rapid separation of cells harboring transcriptionally active vs. inactive proviruses. This reporter enabled us to establish exceptionally large and diverse libraries of hundreds of latently infected T cell lines. Together with human primary cell models, we are using these tools combined with molecular virology, genetics, immunology, and microscopy techniques to dissect HIV-host interactions and understand the mechanisms underlying latency establishment, maintenance, and reversal.
Through this we expect to gain a better understanding of the regulation of HIV-1 latency to get one step closer to achieving a cure for HIV.
Publications
Selected publications:
- Schaefer-Babajew D*, Wang Z*, Muecksch F*, Cho A*, Loewe M, Cipolla M, Raspe R, Johnson B, Canis M, DaSilva J, Ramos V, Turroja M, Millard KG, Schmidt F, Witte L, Dizon J, Shimeliovich I, Yao KH, Oliveira TY, Gazumyan A, Gaebler C, Bieniasz PD, Hatziioannou T, Caskey M, Nussenzweig MC (2023) Antibody feedback regulates immune memory after SARS-CoV-2 mRNA vaccination. Nature. 613(7945):735-742
- Schmidt F*, Muecksch F*, Weisblum Y, Da Silva J, Bednarski E, Cho A, Wang Z, Gaebler C, Caskey M, Nussenzweig MC, Hatziioannou T, Bieniasz PD (2022) Plasma Neutralization of the SARS-CoV-2 Omicron Variant. N Engl J Med. 386(6):599-601
- Muecksch F, Wise H, Templeton K, Batchelor B, Squires M, McCance K, Jarvis L, Malloy K, Furrie E, Richardson C, MacGuire J, Godber I, Burns A, Mavin S, Zhang F, Schmidt F, Bieniasz PD, Jenks S, Hatziioannou T (2022) Longitudinal variation in SARS-CoV-2 antibody levels and emergence of viral variants: a serological analysis. Lancet Microbe. 3(7):e493-e502
- Muecksch F*, Wang Z*, Cho A*, Gaebler C, Ben Tanfous T, DaSilva J, Bednarski E, Ramos V, Zong S, Johnson B, Raspe R, Schaefer-Babajew D, Shimeliovich I, Daga M, Yao KH, Schmidt F, Millard KG, Turroja M, Jankovic M, Oliveira TY, Gazumyan A, Caskey M, Hatziioannou T, Bieniasz PD, Nussenzweig MC (2022) Increased memory B cell potency and breadth after a SARS-CoV-2 mRNA boost. Nature. 607(7917):128-134
- Wang Z*, Muecksch F*, Schaefer-Babajew D*, Finkin S*, Viant C*, Gaebler C*, Hoffmann HH, Barnes CO, Cipolla M, Ramos V, Oliveira TY, Cho A, Schmidt F, Da Silva J, Bednarski E, Aguado L, Yee J, Daga M, Turroja M, Millard KG, Jankovic M, Gazumyan A, Zhao Z, Rice CM, Bieniasz PD, Caskey M, Hatziioannou T, Nussenzweig MC (2021) Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 595(7867):426-431
- Muecksch F*, Weisblum Y*, Barnes CO*, Schmidt F*, Schaefer-Babajew D, Wang Z, JC CL, Flyak AI, DeLaitsch AT, Huey-Tubman KE, Hou S, Schiffer CA, Gaebler C, Da Silva J, Poston D, Finkin S, Cho A, Cipolla M, Oliveira TY, Millard KG, Ramos V, Gazumyan A, Rutkowska M, Caskey M, Nussenzweig MC, Bjorkman PJ, Hatziioannou T, Bieniasz PD (2021) Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity. 54(8):1853-1868 e1857
- Cho A*, Muecksch F*, Schaefer-Babajew D*, Wang Z*, Finkin S*, Gaebler C, Ramos V, Cipolla M, Mendoza P, Agudelo M, Bednarski E, DaSilva J, Shimeliovich I, Dizon J, Daga M, Millard KG, Turroja M, Schmidt F, Zhang F, Tanfous TB, Jankovic M, Oliveria TY, Gazumyan A, Caskey M, Bieniasz PD, Hatziioannou T, Nussenzweig MC (2021) Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature. 600(7889):517-522
- Bou-Nader C*, Muecksch F*, Brown JB, Gordon JM, York A, Peng C, Ghirlando R, Summers MF, Bieniasz PD, Zhang J (2021) HIV-1 matrix-tRNA complex structure reveals basis for host control of Gag localization. Cell Host Microbe. 29(9):1421-1436 e1427
- Robbiani DF*, Gaebler C*, Muecksch F*, Lorenzi JCC*, Wang Z*, Cho A*, Agudelo M*, Barnes CO*, Gazumyan A*, Finkin S*, Hagglof T*, Oliveira TY*, Viant C*, Hurley A, Hoffmann HH, Millard KG, Kost RG, Cipolla M, Gordon K, Bianchini F, Chen ST, Ramos V, Patel R, Dizon J, Shimeliovich I, Mendoza P, Hartweger H, Nogueira L, Pack M, Horowitz J, Schmidt F, Weisblum Y, Michailidis E, Ashbrook AW, Waltari E, Pak JE, Huey-Tubman KE, Koranda N, Hoffman PR, West AP, Jr., Rice CM, Hatziioannou T, Bjorkman PJ, Bieniasz PD, Caskey M, Nussenzweig MC (2020) Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 584(7821):437-442
- Mucksch F*, Citir M*, Luchtenborg C, Glass B, Traynor-Kaplan A, Schultz C, Brugger B, Krausslich HG (2019) Quantification of phosphoinositides reveals strong enrichment of PIP2 in HIV-1 compared to producer cell membranes. Sci Rep. 9(1):17661
- Mucksch F, Laketa V, Muller B, Schultz C, Krausslich HG (2017) Synchronized HIV assembly by tunable PIP2 changes reveals PIP2 requirement for stable Gag anchoring. Elife. 6
Group members
Dr. Frauke Mücksch, Principal Investigator
mail
CIID Room 301, Phone: +49 6221 56-35643
Jannik Löhr, technician
mail
CIID Room 302, Phone: +49 6221 56-1326
Claudia Bastl, PhD student
mail
CIID Room 302, Phone: +49 6221 56-1326
Xinyi Cui, PhD student
mail
CIID Room 302, Phone: +49 6221 56-1326
Luca Menges, PhD student
mail
CIID Room 302, Phone: +49 6221 56-1326
Tetyana Murdza, PhD student
mail
CIID Room 302, Phone: +49 6221 56-1326
Patcharaporn Rathgeber, Admin Assistant
mail
INF 324, Room 406, Phone: +49 6221 56-39747
Group Leader
This group is located on the 3rd floor of the Center for Integrative Infectious Disease Research (CIID) at INF344 and supported by the Chica and Heinz Schaller Foundation Heidelberg.

