Research Group Lusic
Research projects in our group focus on "Nuclear Architecture in Viral Infection".
Projects
We study nuclear architecture and chromatin organization in response to HIV‑1 infection. The nucleus is a complex environment, in which chromatin is organized to support different structural and functional aspects of cell physiology, and represents a challenge for an incoming viral genome, which needs to be integrated into cellular DNA to ensure a productive infection. Integration site selection has functional consequences for viral transcription, which usually follows the integration event. Alternatively, following integration, HIV‑1 can enter the state of latency due to the attenuated or repressed viral transcription. Latency is maintained under current antiviral therapies, but once therapy is interrupted, the virus can resume its productive phase.
Linking HIV‑1 Latency and 3D Genome Organization
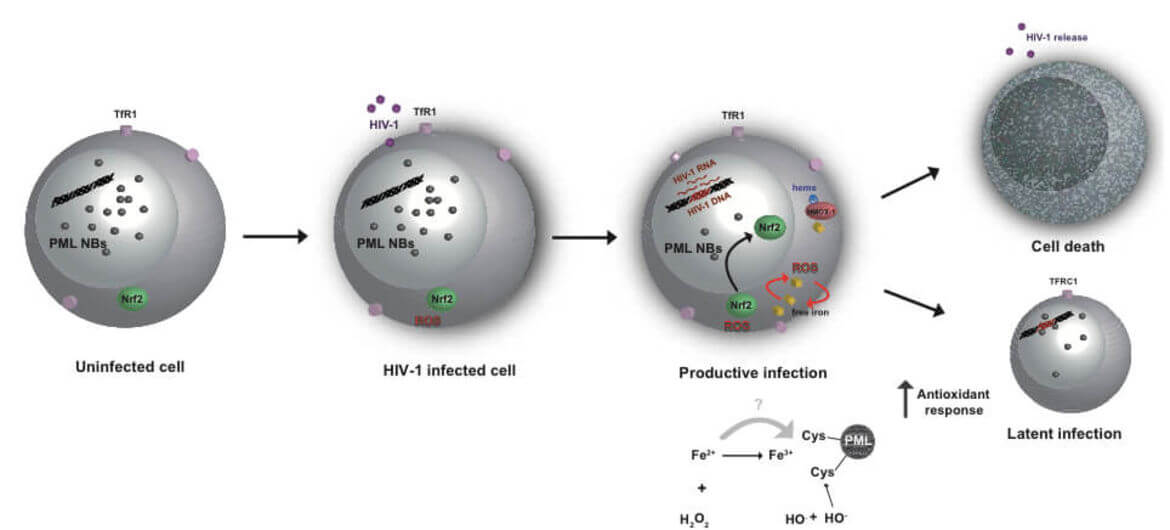
The main obstacle to the functional HIV cure are the latent reservoirs of provirus established in memory CD4+ T cells which cannot be eradicated with current antiviral treatments (cART). Despite some evidence that cART intensification is having positive effects on HIV residual replication, our understanding of HIV‑1 latency and reactivation is still incomplete. While exploring the molecular mechanisms involved in the establishment and maintenance of latency in CD4+ T cells, we obtained substantial evidence that regulation of oxidative phosphorylation pathway and oxidative stress in the cell plays a very important role in HIV‑1 transcriptional control and latency. Our results highlight for the first time the connection and the molecular mechanism between HIV‑1 induced oxidative stress, viral production/latency and Promyelocytic Leukemia Nuclear Bodies (PML NBs) turnover and further strengthen the idea that PML NBs can be used both as molecular markers and/or pharmacologic targets for HIV‑1 infection.
Selected Publications
- Fronza, R., Lusic, M., Schmidt, M. and Lucic, B. (2020). Spatial–Temporal Variations in Atmospheric Factors Contribute to SARS-CoV‑2 Outbreak. Viruses 12(6), 588.
- Shytaj, I.L., Lucic, B., Forcato, M., Penzo, C., Billingsley, J., Laketa, V., Bosinger, S., Stanic, M., Gregoretti, F., Antonelli, L., Oliva, G., Frese C. K., Trifunovic, A., Galy, B., Eibl, C., Silvestri, G., Bicciato, S., Savarino A. and Lusic M. (2020). Alterations of redox and iron metabolism accompany the development of HIV latency. EMBO J. e102209.
- Lucic B, Chen H‑C, Kuzman M, Zorita E„ Wegner J, Minneker V, Wang W, FronzaR, Laufs S, Schmidt M, Stadhouders R, Roukos V, Vlahovicek K, Filion GJ and Lusic M. Spatially clustered loci with multiple enhancers are frequent targets of HIV‑1 Nature Com Accepted for publication. IF 12,353
- Muenchau S, Deutsch R, de Castro IJ, Hielscher T, Heber N, Niesler B, Lusic M, Stanifer ML, Boulant S. (2019) Hypoxic environment promotes barrier formation in human intestina epithelial cells through regulation of miRNA-320a expression. Mol Cell Biol. IF 3.813
- Ali H, Mano M, Braga L, Naseem A, Marini B, Vu DM, Collesi C, Meroni G, Lusic M, and Giacca, M.(2019) Cellular TRIM33 restrains HIV‑1 infection by targeting viral integrase for proteasomal degradation. Nat Commun, 10:926. IF 12,353
- Michieleto D, Lusic M, Orlandini E, Marenduzzo D (2019) Physical principles of retroviral integration. Nat Commun, 10:575. IF 12,353
- Bejarano DA, Peng K, Laketa V, Börner K, Jost KL, Lucic B, Glass B, Lusic M, Müller B, Kräusslich HG. (2019) HIV‑1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex. Elife. IF 7,616
- Lusic M. (2018) Coloring Hidden Viruses. Elife. IF 7,616
- Agostini S, Ali H, Vardabasso C, Fittipaldi A, Tasciotti E, Cereseto A, Bugatti A, Rusnati M, Lusic M, Giacca M (2017) Inhibition of Non Canonical HIV‑1 Tat Secretion Through the Cellular Na+,K+-ATPase Blocks HIV‑1 Infection. EBioMedicine. 21:170–181. IF 6.183
- Lusic M and Robert F. Siliciano (2017) Nuclear landscape of HIV‑1 infection and integration. Nat Rev Microb. 15:69–82. IF 31,851
- Lucic B and Lusic M. (2016) Connecting HIV‑1 integration and transcription: a step toward new treatments. FEBS Letters. 590:1927. IF 2,999
- Turrini F, Marelli S, Kajaste-Rudnitski A, Lusic M, Van Lint C, Das AT, Harwig A, Berkhout B, Vicenzi E. (2015) HIV transcriptional silencing caused by TRIM22 inhibition of Sp1 binding to the viral promoter Retrovirology, 12:104. IF 3,867
- Marini B, Kertesz-Farkas A, Lucic B, Hashim A, Lisek K, Manganaro L, Pongor S, Luzzati R, Mavilio F, Giacca M and Lusic M (2015) Nuclear architecture dictates HIV‑1 integration site selection Nature, 521: 227–31. IF 41.577
- Lusic M and Giacca M (2014) Ground Control to Major Tom: “Prepare for HIV Landing” Cell Host & Microbe, 16: 557–559. IF 17,872
- Lusic M and Giacca M (2014) Regulation of HIV‑1 latency by chromatin structure and nuclear architecture. A review article for the special issue “Functional Relevance and Dynamics of Nuclear Organization. J Mol Biol. IF 4.894
- Felician G, Collesi C, Lusic M, Martinelli V, Ferro MD, Zentilin L, Zacchigna S, Giacca M (2014) Epigenetic modification at Notch responsive promoters blunts efficacy of inducing notch pathway reactivation after myocardial infarction. Circ Res, 115:636–49. IF 15,211
- Lusic M, Marini B, Ali H, Lucic B, Luzzati R, and Giacca M (2013) Proximity to PML Nuclear Bodies negatively regulates HIV‑1 gene expression in CD4+ T cells. Cell Host & Microbe, 13: 665–677. Research highlight in Science Vol 341 (2013) and in Cell Host & Microbe 13:625–626. IF 17,872
- Della Chiara G, Crotti A, Liboi E, Giacca M, Poli G and Lusic M. (2011) Negative Regulation of HIV- 1 Transcription by a Heterodimeric NF-kB1/p50 and C‑Terminally Truncated STAT5 Complex. J Mol Biol, 410: 933–943. IF 4,894
- Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M and Cereseto A (2011) KAP‑1 inhibits HIV‑1 integration. Cell Host & Microbe, 9: 484–95. IF 17,872
- Manganaro L, Lusic M*, Gutierrez MI, Cereseto A, Del Sal G and Giacca M* (2010) Concerted action of cellular JNK and Pin‑1 restricts HIV‑1 genome integration to activated CD4+ T lymphocytes. Nat Medicine, 16: 329–323 (* corresponding authors). IF 32,621
- Dieudonné M, Maiuri P, Biancotto C, Knezevich A, Kula A,Lusic M, and Marcello A. (2009) Transcriptional competence of the integrated HIV‑1 provirus at the nuclear periphery EMBO J. 28:2231–2243. IF 9,792
- Sabo’ A, Lusic M, Cereseto A and Giacca M (2008) Acetylation of conserved lysines in the catalytic core of CDK9 regulates kinase activity and subnuclear localization. Mol Cell Biol, 28:2201–12 . IF 3,813
- Vardabasso C, Manganaro L, Lusic M, Marcello A and Giacca M (2008) The histone chaperone protein Nucleosome Assembly Protein‑1 (hNAP‑1) binds HIV‑1 Tat and promotes viral transcription. Retrovirology, 28:5–8. IF 3,867
- Perkins KJ, Lusic M, Mitar I, Giacca M and Proudfoot NJ (2008) Transcription dependent gene looping of the HIV‑1 provirus is dictated by recognition of pre-mRNA processing signals. Molecular Cell, 29:56–68. IF 14,248
Group Members
Dr. Marina Lusic, Principal Investigator
Marina.Lusic(at)med.uni-heidelberg.de
CIID Room 204a, +49 6221 56-5007
Sara Giardina, PhD Student
Sara.Giardina(at)med.uni-heidelberg.de
Dr. Bojana Lucic, Senior Postdoc
Bojana.Lucic(at)med.uni-heidelberg.de
CIID Room 201, +49 6221 56-7874
Dr. Carlotta Penzo, Postdoc
Carlotta.Penzo(at)med.uni-heidelberg.de
CIID Room 224, +49 6221 56-4128
Nadine Weissheimer, Technician
Nadine.Weissheimer(at)med.uni-heidelberg.de
Alumni
Former Lab Members
- Ankit Arora
- Julia Heinze
- Martin Kappmann
- Mia Stanic

