Research Group Chlanda
Research projects in our group focus on "Cryo-Electron Microscopy of Viral Infection".
Projects
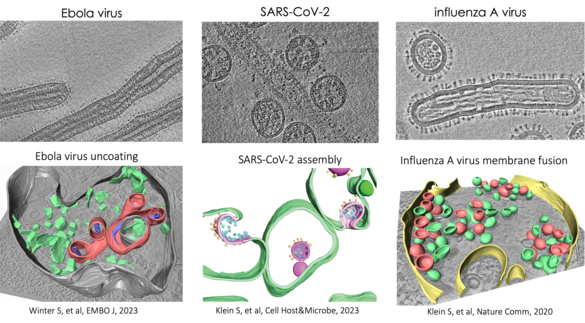
We study membrane-enveloped RNA viruses, including influenza A virus and other members of the Orthomyxoviridae family, as well as SARS-CoV-2 and Ebola virus. Our goal is to understand the molecular mechanisms of key steps involving virus-host membrane interactions during the viral infection cycle. Our research focuses on virus-mediated membrane fusion, the structure and function of replication organelles, and virus assembly.
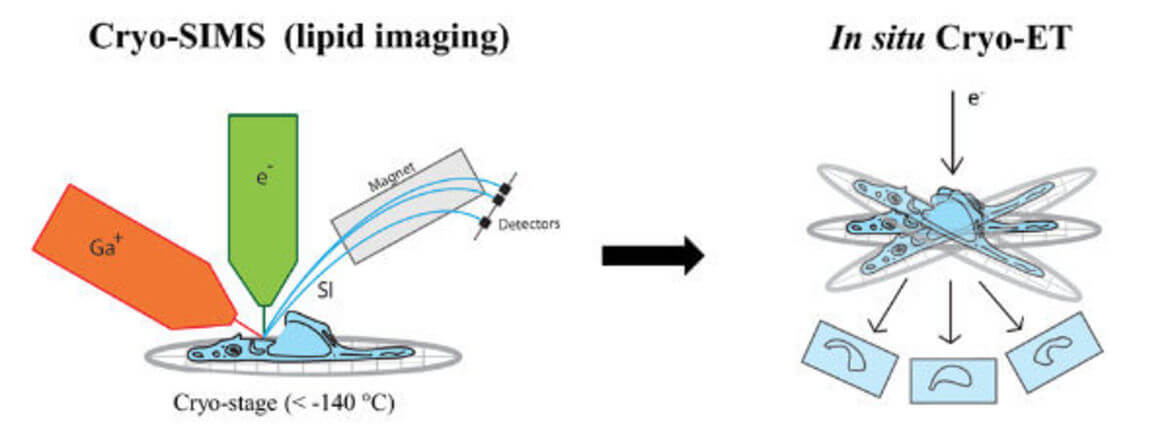
These critical processes of viral infection remain poorly understood at high resolution in the context of infected cells. To address this, we use and further develop cryo-electron microscopy (cryo-EM) techniques such as cryo-focused ion beam milling (cryo-FIB), in situ cryo-electron tomography (cryo-ET), and cryo-correlative light and electron microscopy (cryo-CLEM). We also integrate other imaging methods, including fluorescence microscopy and cryogenic imaging mass spectrometry, to unravel the complexities of viral-membrane interactions.
Entry and Spread of Influenza A Virus
We specifically investigate hemagglutinin-mediated membrane fusion and the entry of spherical and filamentous influenza A viruses. We have developed a correlative fluorescence and scanning electron microscopy approach that allows us to assess virus morphology changes during viral spread. Additionally, we examine how neutralizing antibodies and mucins influence the spread of viruses with different morphologies. This project is funded by SFB1129.
Assembly and vRNP Trafficking in Orthomyxoviridae
We discovered that influenza A virus vRNPs interact with hemagglutinin-remodeled membranes, facilitating vRNP clustering. We are currently investigating vRNP incorporation into budding virions, orchestrated by M1 and M2 proteins, in both non-polarized and polarized cell lines using different members of the Orthomyxoviridae family.
SARS-CoV-2 Replication Organelles and Assembly of Virus-Like Particles
Using cryo-ET, we study SARS-CoV-2 assembly in a virus-like particle system. We aim to understand how assembly is orchestrated at the molecular level and the roles of individual proteins in membrane bending. Additionally, we focus on the structural characterization of SARS-CoV-2 double-membrane vesicles and a molecular pore.
Spatial Lipidomics and Viral Infection
Secondary ion mass spectrometry (SIMS) allows non-invasive imaging of chemically unmodified lipids with high chemical specificity. However, SIMS has so far been only performed on chemically fixed and dehydrated samples (sample preparation procedures known to severely alter membrane structure). In collaboration with Tom Wirtz (Luxembourg Institute of Science and Technology), we will apply SIMS to vitreous cryo-lamellas of the cells prepared by focused-ion beam milling to provide a spatial map of lipids in cellular organelles at native conditions. The lipid map will be subsequently correlated to the membrane structures observed by cryo-ET. We study lipids such as cholesterol which is crucial in host-pathogen interactions as well as in membrane trafficking.
Selected Publications
Complete Publication List (PubMed)
- Nucleocapsid assembly drives Ebola viral factory maturation and dispersion.
Vallbracht M, Bodmer BS, Fischer K, Makroczyova J, Winter SL, Wendt L, Wachsmuth-Melm M, Hoenen T, Chlanda P. Cell. 2025 Feb 6;188(3):704-720.e17. doi: 10.1016/j.cell.2024.11.024. Epub 2024 Dec 31.PMID: 39742805 - Winter SL, Chlanda P, (2023) The Art of Viral Membrane Fusion and Penetration, Subcell Biochem, Book: Virus Infected cells. doi: 10.1007/978–3‑031–40086-5_4.
- Zimmermann L, Zhao X, Makroczyova J, Wachsmuth-Melm W, Prasad V, Hensel Z, Bartenschlager R, Chlanda P. (2023) SARS-CoV‑2 nsp3 and nsp4 are minimal constituents of a pore spanning replication organelle. Nature Communications,14(1):7894. doi: 10.1038/s41467-023–43666‑5
- Zimmermann L, Chlanda P. Cryo-electron tomography of viral infection — from applications to biosafety. Curr Opin Virol. 2023 Aug;61:101338. doi: 10.1016/j.coviro.2023.101338.
- Winter SL, Golani G, Lolicato F, Vallbracht M, Thiyagarajah K, Ahmed SS, Lüchtenborg C, Fackler OT, Brügger B, Hoenen T, Nickel W, Schwarz US, Chlanda P, (2023) The Ebola virus VP40 matrix undergoes endosomal disassembly essential for membrane fusion, EMBO J. 42(11):e113578. doi: 10.15252/embj.2023113578.
- Klein S, Golani G, Lolicato F, Beyer D, Herrmann A, Wachsmuth-Melm M, Reddmann N, Brecht R, Lahr C, Hosseinzadeh M, Kolovou A, Schorb M, Schwab Y, Brügger B, Nickel W, Schwarz US, Chlanda P, (2023) IFITM3 blocks viral entry by sorting lipids and stabilizing hemifusion, Cell Host&Microbe, S1931-3128(23)0019–9, doi: 10.1016/j.chom.2023.03.005.
- Winter SL, Chlanda P, (2021) Dual-axis Volta phase plate cryo-electron tomography of Ebola virus-like particles reveals actin-VP40 interactions, J Struct Biol, 107742. doi: 10.1016/j.jsb.2021.107742.
- Klein S, Wimmer WH, Winter SL, Kolovou A, Laketa V, Chlanda P, (2021) Post-correlation on-lamella cryo-CLEM reveals the membrane architecture of lamellar bodies. Communications Biology, 2021 Jan 29;4(1):137. doi: 10.1038/s42003-020–01567‑z.
- Klein S, Wachsmuth-Melm M, Winter SL, Kolovou A, Chlanda P. (2021) Cryo-correlative light and electron microscopy workflow for cryo-focused ion beam milled adherent cells. Methods Mol Biol. Correlative Light and Electron Microscopy, IV Volume 162, Chapter 12.
- Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P. (2020) SARS-CoV‑2 structure and replication characterized by in situ cryo-electron tomography. Nature Communications, 11, 5885 doi.org/10.1038/s41467-020–19619‑7.
- Klein S, Müller TG, Khalid D, Sonntag-Buck V, Heuser AM, Glass B, Meurer M, Morales I, Schillak A, Freistaedter A, Ambiel I, Winter SL, Zimmermann L, Naumoska T, Bubeck F, Kirrmaier D, Ullrich S, Barreto Miranda I, Anders S, Grimm D, Schnitzler P, Knop M, Kräusslich HG, Dao Thi VL, Börner K, Chlanda P. (2020), SARS-CoV‑2 RNA Extraction Using Magnetic Beads for Rapid Large-Scale Testing by RT-qPCR and RT-LAMP. Viruses. 12(8):863. doi: 10.3390/v12080863.
Group Members
- Prof. Dr. Petr Chlanda, Principal Investigator
Petr.Chlanda(at)bioquant.uni-heidelberg.de
BioQuant Room 102a, +49 6221 54-51231
- Raffael Balkeny, PhD Student
raffael.balkeny(at)bioquant.uni-heidelberg.de
BioQuant Room 102, +49 6221 54-51230
- Konstantin Fischer, PhD Student
konstantin.fischer(at)bioquant.uni-heidelberg.de
BioQuant Room 103a, +49 6221 54-51230
- Dr. Tim Krischuns, Postdoc
tim.krischuns(at)bioquant.uni-heidelberg.de
BioQuant Room 103, +49 6221 54-51230
- Dr. Jana Makroczyová, Lab Manager
Jana.Makroczyova(at)bioquant.uni-heidelberg.de
BioQuant Room 102, +49 6221 54-51230
- Roberta Malamud, MSc Student
roberta.malamud(at)stud.uni-heidelberg.de
BioQuant Room 161, +49 6221 54-51251
- Sarah Peterl, PhD Student
sarah.peterl(at)bioquant.uni-heidelberg.de
BioQuant Room 103, +49 6221 54-51230
- Varvara Plotnikova, PhD Student
varvara.plotnikova(at)bioquant.uni-heidelberg.de
BioQuant Room 161, +49 6221 54-51251
- Patcharaporn Rathgeber, Administrative Assistant
Patcharaporn.Rathgeber@med.uni-heidelberg.de
INF 324 Room 406, +49 6221 56-39747
- Alexandra Reimers, MSc Student
A.Reimers(at)stud.uni-heidelberg.de
BioQuant Room 161, +49 6221 54-51251
- Dr. Moritz Wachsmuth-Melm, Postdoc
Wachsmuth-Melm(at)stud.uni-heidelberg.de
BioQuant Room 103a /161, +49 6221 54-51251
- Dr. Liv Zimmermann, Postdoc
Liv.Zimmermann(at)bioquant.uni-heidelberg.de
BioQuant Room 103a /161, +49 6221 54-51251
Alumni
Former Lab Members
- Lisa Augenstein, M.Sc. Student
- Konstantin Fischer, M.Sc. Student
- Steffen Klein, PhD Student
- Androniki Kolovou, Technical Assistant
- Carmen Lahr, M.Sc. Student
- Anna Michalczyk-Schneider, Administrative Assistant
- Carl Niklas Schneider, M.Sc. Student
- Keerthihan Thiyagarajah, M.Sc. Student
- Melina Vallbracht, Postdoc
- Benedikt Wimmer, M.Sc. Student
- Sophie Winter, PhD Student
- Xiaohan Zhao, M.Sc. Student
Group Leader
For more information about the Chlanda Lab, please check out Website Chlanda Lab
This group is located on the 1st floor of the Center for Quantitative Analysis of Molecular and Cellular Biosystems (BioQuant) at INF 267 and supported by the Chica and Heinz Schaller Foundation Heidelberg