Research Group Dalpke
Group Leader: Prof. Dr. med Alexander Dalpke
RESEARCH INTERESTS
The focus of my research is infection and immunity. Current work centers on two topics:
1. We are studying the recognition of microbial nucleic acids by pattern recognition receptors. We try to elucidate principles that enable Toll-like receptors to differentiate foreign, microbial from self nucleic acids. We address the importance of RNA modifications on TLR activation, specifically focusing on 2’-O-methyl modifications. Moreover we studyand the importance of RNA processing on immune stimulation with running projects on RNase 6 and 7. Finally,we are also interested in longterm effects of bacterial encounter on innate immune cells, a condition that has been termed trained immunity.
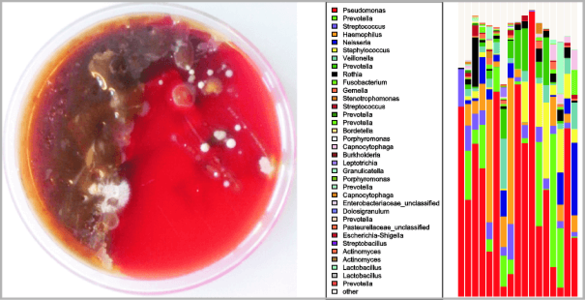
2. A second project area studies polymicrobial infections by means of next generation sequencing. We focus on patients suffering from cystic fibrosis (CF) but also use the technique to explore microbiome changes in other diseases. In CF we aim to analyze microbiome alterations in early childhood as well as during exacerbation and focus on interactions of commensal and pathogenic bacteria. Specifically we try to elucidate to what extent and how commensal bacteria affect growth and inflammation by classical pathogens. Currently we are focusing on the effects of short chain fatty acids.
Funding: DFG,, TRR156, TRR319, Mukoviszidose e.V.
Immunostimulation by nucleic acids
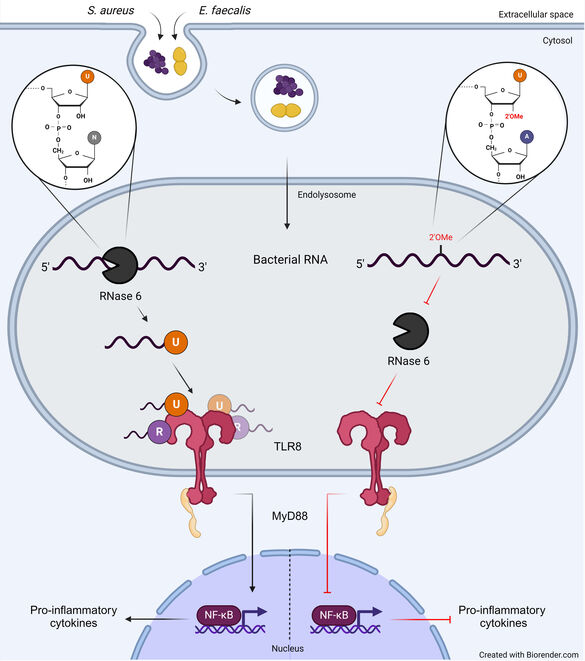
Cells of the innate immune system recognize conserved microbial structures, so called pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors. Among these receptors Toll-like receptors (TLR) 3, -7, -8, -9 and -13 recognize nucleic acids of viral, bacterial or synthetic origin. However, the exact mechanisms of nucleic acid recognition and the differentiation between foreign (dangerous) and self (usually harmless) are still only known partially. Work of our own could show (1) that bacterial RNA represents a PAMP recognized by various immune cells. We could also show that TLR13 is a murine receptor for bacterial RNA. (2) We work onposttranscriptional RNA modifications and their role role for self/non-self discrimination. We identified 2’-O-methylation as a modification that inhibits TLR7 and 8 stimulation. This naturally occurring RNA modification can be regulated under stress within bacteria thus potentially contributing to immune evasion. (3) Recent projects analyze the role of RNases in the process of TLR8 stimulation, as the receptor is activated by RNA breakdown products instead of full length RNA. We show that RNA processing by RNase6 is a prerequisite and critical for RNA-TLR interactions in monocytes. Of note, RNA modifications and RNA degradation act together. An ongoing project addresses the importance of RNase 7 for RNA recognition in the interplay of skin cells and monocytes.
Selected publications
Nunes IV*, Breitenbach L*, Pawusch S, Eigenbrod T, Ananth S, Schad P, Fackler OT, Butter F, Dalpke AH, Chen LS (2024): Bacterial RNA sensing by TLR8 requires RNase 6 processing and is inhibited by RNA 2’O-methylation. EMBO Rep, doi.org/10.1038/s44319-024-00281
Galvanin A, Vogt LM, Grober A, Freund I, Ayadi L, Bourguignon-Igel V, Bessler L, Jacob D, Eigenbrod T, Marchand V, Dalpke A, Helm M, Motorin Y (2020): Bacterial tRNA 2′-O-methylation is dynamically regulated under stress conditions and modulates innate immune response. Nucl Acid Res, 48(22): 12833-12844. doi: 10.1093/nar/gkaa1123
Hafner A, Kolbe U, Freund I, Castiglia V, Kovarik P, Poth P, Herster F, Weigand MA, Weber AN, Dalpke AH and Eigenbrod T (2019): Crucial role of nucleic acid sensing via endosomal Toll-like receptors for the defense of Streptococcus pyogenes in vitro and in vivo. Front Immunol, 10:198.doi: 10.3389/fimmu.2019.00198
Eigenbrod T and Dalpke AH (2015): Bacterial RNA: An Underestimated Stimulus for Innate Immune Responses. J Immunol 195(2): 411-8
Eigenbrod T, Pelka K, Latz E, Kreikemeyer B and Dalpke AH (2015): TLR8 Senses Bacterial RNA in Human Monocytes and Plays a Nonredundant Role for Recognition of Streptococcus pyogenes. J Immunol. 195(3): 1092-1099, doi: 10.4049/jimmunol.1403173
Hidmark A, von Saint Paul A, Dalpke AH (2012): Cutting Edge: TLR13 is a receptor for bacterial RNA. J Immunol. 189(6):2717-21
Gehrig S, Eberle ME, Botschen F, Rimbach K, Eberle F, Eigenbrod T, Kaiser S, Holmes WM, Erdmann VA, Sprinzl M, Bec G, Keith G, Dalpke AH* and Helm M* (2012): Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J Exp Med 209 (2): 225-233, doi 10.1084/jem.20111044
Microbiome analysis in cystic fibrosis
Interactions between bacteria and their host represent a full continuum from pathogenicity to mutualism. From an evolutionary perspective, host-bacteria relationships are no longer considered a two-component ecological system but rather a complex interactive network. Cystic fibrosis (CF) is characterized by defective mucociliary clearance associated with polymicrobial chronic airway infections. Those infections, leading to persistent inflammation and periodic episodes of acute pulmonary exacerbation, contribute to an irreversible decline in CF lung function. However, the link between bacterial infections and decline in CF lung function is not yet understood. We study CF lung microbiome aiming to answer different questions about CF microbiome resilience, antibiotic treatment effect and possible dysbiosis induced by exacerbation. In a runningproject we address the hypothesis that certain airway commensal bacteria might interfere with CF pathogens and thus exert rather beneficial effects. This work focuses on interactions of commensal bacteria with Pseudomonas aeruginosa within in vitro systems as well as microbiome and metagenomics studies from patients. Specifically we focus on short chain fatty acids produced by commensals and how these affect growth of pathogenic bacteria as well as inflammation. The projects combines in vitro work with a clinical study.
Selected publications
Tony-Odigie A, Dalpke1 AH, Boutin S, Yi B (2024): Airway commensal bacteria in cystic fibrosis inhibit the growth of P. aeruginosa via a released metabolite. Microbiol. Res., 283:127680, https://doi.org/10.1016/j.micres.2024.127680
Frey DL, Bridson C, Dittrich S, Graeber SY, Stahl M, Wege S, Herth F, Sommerburg O, Schultz C, Dalpke A*, Mall M*, Boutin S* (2022): Changes in microbiome dominance are associated with declining lung function and fluctuating inflammation in people with Cystic Fibrosis. Front Microbiol, 13:885822, doi:10.3389/fmicb.2022.885822
Tony-Odigie A, Wilke L, Boutin S, Dalpke AH and Yi B (2022): Commensal bacteria in the cystic fibrosis airway microbiome reduce P. aeruginosa induced inflammation. Front Cell Infect Microbiol 12:824101, doi: 10.3389/fcimb.2022.824101
Metzger MI, Gräber SY, Stahl M, Sommerburg O, Mall MA, Dalpke AH and Boutin S (2021): A volatile and dynamic longitudinal microbiome is associated with less reduction in lung function in adolescents with cystic fibrosis. Front Cell Infect Microbiol, 11:763121. doi: 10.3389/fcimb.2021.763121
Frey DL*, Boutin S*, Dittrich S*, Graeber SY, Stahl M, Wege S, Herth F, Sommerburg O, Schultz C, Mall M, Dalpke A (2021): Relationship between airway dysbiosis, inflammation and lung function in adults with cystic fibrosis. J Cyst Fibr, S1569-1993(20):30954-1
Kolbe U, Yi B, Poth T, Saunders A, Boutin S, Dalpke A (2020): Early cytokine induction upon Pseudomonas aeruginosa infection in murine precision cut lung slices depends on sensing of bacterial viability. Front Immunol, 11: 598636. doi: 10.3389/fimmu.2020.598636
Boutin S, Graeber SY, Stahl M, Dittrich SA, Mall MA and Dalpke AH (2017): Chronic but not intermittent infection with Pseudomonas aeruginosa is associated with global changes of the lung microbiome in cystic fibrosis. Eur Respir J 50 (4): 1701086, doi: 10.1183/13993003.01086-2017
Supported by Mukoviszidose e.V.:
Links:
Mukoviszidose e.V.
https://www.muko.info/angebote/forschungsfoerderung/gefoerderte-projekte/#c2963
link to blog
Curriculum Vitae
| Curriculum Vitae | |
| Since 2023 | PI, TRR156 „The skin as sensor and effector organ orchestrating local and systemic immunity”, project B05 |
| Since 2022 | Full Professor (W3) for Medical Microbiology and Hygiene, Medical Director, Medical Microbiology and Hygiene, Dept. of Infectious Diseases, University Hospital Heidelberg |
| Since 2021 | PI, TRR319 “RMaP, RNA modifications and processing”, project A3 |
| 2019-2022 | Director, Institute of Med. Microbiology and Hygiene, Institute of Virology, Med. Faculty, Technical University Dresden |
| 2015-2018 | Chairman of the Habilitation Committee I, Medical Faculty Heidelberg |
| 2014-2018 | PI, German Center for Lung Research (DZL), TLRC Heidelberg |
| 2012-2015 | Coordinator Teaching & Education Activities, DZIF, Heidelberg site |
| 2011 | Consultant in microbiology, virology and epidemiology of infections |
| 2011-2015 | Member of SFB 938 “Environment specific control of immunological reactivity"; project E |
| 2008-2011 | Speaker of the Postgraduate Program "Differential activation and integration of signaling modules within the immune system"; postgraduate program of Baden-Württemberg |
| 2007-2018 | Member of the "Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology" |
| 2006-2009 | Member of the SFB405 "Immunotolerance and its disturbances"; project B18 |
| 2006-2018 | Professor for Medical Microbiology and Infection and Immunity (W3), Dept. of Medical Microbiology and Hygiene, University Heidelberg |
| 2006 | Specialist in immunology ("Fachimmunologe DGfl", German Society for Immunology |
| 2005-2006 | Group Leader, Dept. of Medical Microbiology and Hygiene, University Heidelberg |
| 2004-2008 | Member of the DFG priority programm SP1110 "Innate Immunity" |
| 2004 | Habilitation, university lecturer for infection and immunity Topic: Suppressor of Cytokine Signaling (SOCS) proteins as regulators of the activity of the innate immune system |
| 1999-2004 | Post-doc and Research Assistant, Inst. of Medical Microbiology, Philipps-University Marburg |
| 1998-1999 | First-year resident, Kreis- und Stadtkrankenhaus Alfeld, Internal medicine, License to practice medicine („Approbation als Arzt“) |
| 1992- 1998 | Medical student, Georg-August University Göttingen |
Lab members
My group just recently moved to Heidelberg. Positions available and applications welcome!
| Name | Function | Phone +49 6221 56- |
| Dr. Lan-Sun Chen | postdoc | 34783 |
| Andrew Tony-Odigie | postdoc | 310172 |
| Ivaneiá Valeriano-Nunes | PhD Student | |
| Luisa Breitenbach | PhD Student | |
| Paulina Schad | PhD Student | |
| Mai Wang | PhD Student | |
| Suzan Jakob | technician | 36094 |