AG Bartenschlager

Ralf Bartenschlager
Head of the Department
"MOLECULAR VIROLOGY"
Phone: +49 (0)6221-56 4225
Fax: +49 (0)6221-56 4570
Our original interests focused on the molecular and cellular biology underlying the HCV replication cycle. In the last few years, we expanded our scope and included other medically relevant viral pathogens: flaviviruses such as Dengue virus and Zika virus, hepatitis B and D virus and, very recently, SARS-CoV-2.
Our studies revolve around virus – host cell interaction using as major technique novel imaging methods and high resolution microscopy. Moreover, we are actively working on innate immune responses induced by infections and viral countermeasures. Finally, we aim to develop antiviral drugs and to characterize novel drugs for their mode-of-action.
Research Team Members















Research Projects
The research areas described below apply to HCV, DENV and ZIKV as well as SARS-CoV-2 that we study in parallel in order to identify principles that are either unique to each virus or that might apply to viruses more general.
I. BIOGENESIS AND 3D STRUCTURE OF MEMBRANOUS REPLICATION ORGANELLES OF RNA VIRUSES
Replication of plus-strand RNA viruses occurs in close association with intracellular membranes. We have earlier shown that Dengue virus (DENV) induces invaginations of ER membranes (Welsch et al., Cell Host & Microbe 2009) whereas HCV induces double-membrane vesicles that appear as protrusions from the ER (Romero-Brey et al., Plos Pathogens 2012). Moreover, we found that SARS-CoV-2 induces double-membrane vesicles having morphology similar to the ones induced by HCV (Cortese et al., Cell Host & Microbe 2020).
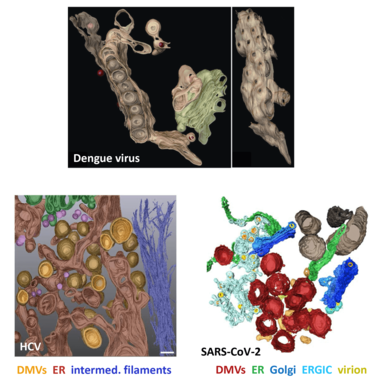
By using a combination of light and electron microscopy techniques we study the 3D architecture of these virus-induced membrane alterations and try to identify functional sub-compartments. We combine these data with functional and mechanistic analyses, including proteome and lipidome analyses, in order to understand the cell biology underlying the biogenesis of these membranous replication organelles.
Finally, we aim to establish in vitro reconstitution systems allowing to understand the biophysics underlying the induction of membrane curvature and alterations of lipid compositions of viral replication organelles.
In all cases, the organelles are derived from the ER. DMVs, double-membrane vesicles; ERGIC, ER-Golgi intermediate compartment; intermed. filaments, intermediate filaments. Panels after Welsch et al., Cell Host Microbe 2008 (Dengue virus); Romero-Brey et al., Plos Pathogens 2012 (HCV); Cortese et al., Cell Host Microbe 2020 (SARS-CoV-2).
By using a combination of light and electron microscopy techniques we study the 3D architecture of these virus-induced membrane alterations and try to identify functional sub-compartments. We combine these data with functional and mechanistic analyses, including proteome and lipidome analyses, in order to understand the cell biology underlying the biogenesis of these membranous replication organelles.
Finally, we aim to establish in vitro reconstitution systems allowing to understand the biophysics underlying the induction of membrane curvature and alterations of lipid compositions of viral replication organelles.
II. Identification of host cell factors regulating the viral replication cycle
By using high throughput screens we want to identify host cell dependency or restriction factors. We use either whole virus-based genome-wide screens or distinct subscreens such as the human kinome (see e.g. Reiss, Rebhan et al., Cell Host & Microbe 2011; Cortese et al., Cell Reports 2019). Hit candidates are validated using approaches described above (section I).
III. Virus assembly
Assembly of virus particles requires the concerted action of viral and host cell proteins as well as distinct lipids. We have earlier shown that HCV assembly is tightly linked to lipid droplets and proposed several mechanisms how infectious virus particles might form (reviewed in Bartenschlager et al., Trends in Microbiology, 2011). In addition, we have determined the lipidome of HCV (Merz, Long et al., Journal of Biological Chemistry, 2011). Building on these results we want to determine how the different steps of the assembly process are coordinated, which cellular proteins are required for that and how virus particles are released from the cell. For this we employ high resolution microscopy methods including electron tomography and live cell imaging. By using this approach, we have for instance found that virus-induced DMVs are tightly associated with lipid droplets (Figure 2). This organization would directly link a major cellular lipid source with HCV replication organelle (DMV) formation and the assembly of highly lipidated virus particles.
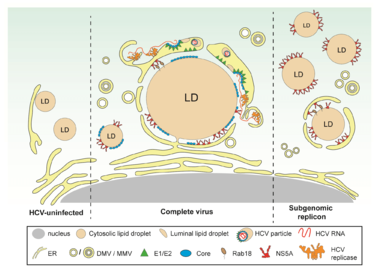
In naive cells (left part of the figure), LDs are either completely cytosolic or peripherally associated with the ER. In HCV-infected cells (middle part of the figure), ER membranes containing structural proteins and replicase are wrapped around LDs. These ER membranes are interconnected with DMVs, the assumed HCV replication site. This close proximity could facilitate virion assembly occurring by budding into the ER lumen. In cells containing a subgenomic replicon capable of self-replication but unable to form virus particles (right part of the figure), NS5A accumulates on the surface of LDs, but only a small fraction of LDs is wrapped by ER membranes.
IV. Innate immune responses against viral infection and viral countermeasures
Virus infections are sensed by pattern recognition receptors leading to, amongst others, the expression of interferons. Viruses in turn have developed strategies to cope with this innate antiviral response. We have earlier shown that HCV blocks signalling pathways by cleaving the adaptor protein MAVS (Meylan et al., Nature 2005) and obtained evidence that HCV persistence might be facilitated by the stochastic nature of the interferon response (Bauhofer et al., Gastroenterology 2012).
We now want to characterize the mechanisms how virus infection triggers an antiviral cytokine response and the mechanism of Interferon-stimulated genes (Metz et al., Hepatology 2012). In these respects, we expanded the scope of our studies to include the hepatitis B and D viruses.
V. Development of antiviral drugs, broad-spectrum antivirals and drug resistance assays
In the case of HCV, numerous direct antiviral drugs have been developed that allow curing more than 95% of treated patients. However, for many other highly prevalent viruses such as DENV and ZIKV no such drugs are available. Therefore, we aim to develop broad-spectrum antiviral drugs, which are of high importance especially for ‘emerging’ viruses, by using a host-targeting approach or nucleos(t)ides (Kaufmann et al., Nature Reviews Drug Discovery, 2018). Moreover, we are studying the mode-of-action of those drugs for which the antiviral mechanism is not known (e.g. Berger et al., Gastroenterology 2014; Kaptein et al., Nature 2021; Riva et al., Journal of Virology 2021).