Research Group Stephanie Roessler
Hepatobiliary carcinogenesis
Liver cancer is the third most common cause of cancer-related death worldwide. Hepatocellular carcinomas (HCC) and biliary tract cancers (BTC) are very heterogenous cancer entities. BTC are classified based on the anatomical location into intrahepatic cholangiocarcinoma (iCCA), perhilar cholangiocarcinoma (pCCA), distal cholangiocarcinoma (dCCA) and gallbladder cancer (GBC). The high mortality rate of HCC and BTC is mainly caused by inflammation, fibrosis, cirrhosis and resistance to chemotherapy. One of the key pathways is the IL-6 signaling pathway which facilitates the interplay between infiltrating immune cells and tumor cells in the tumor microenvironment of the liver. Although significant progress has been made over the last years, the molecular mechanisms and signaling pathways underlying HCC and CCA development and progression are still poorly understood. To better understand the development and progression of liver cancers, we are using cohort-based multi omics approaches, cell culture-based analyses and animal experiments.
Tumor suppressor genes and the tumor microenvironment
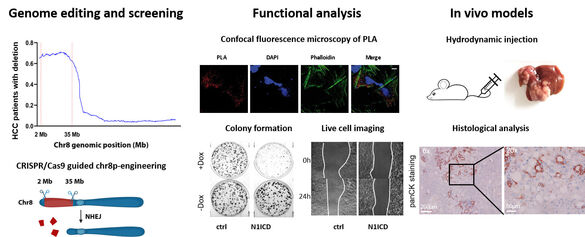
We discovered that chromosome 8p, which exhibits the highest frequency of genomic copy number loss (45%), is linked to poor prognosis of patients with HCC (Roessler et al. Gastroenterology, 2012). In addition, we identified the genes SORBS3 and SH2D4A, located on chromosome 8p, as tumor suppressors that inhibit IL-6 signal transduction (Ploeger et al. Hepatology, 2016). SH2D4A interacted with STAT3 and PHB1 in the mitochondria, its knockdown led to increased mitochondrial respiration, which was reversed by the PHB ligand FL3 (Ploeger, Huth et al. Cell Death Dis, 2020). Using genome-wide CRISPR/Cas9 screening and engineered isogenic cells, we demonstrated that deletion of chromosome 8p sensitizes tumor cells to targeting of the reactive oxygen sanitizing enzyme NUDT17 (Huth et al. Science Advances, 2023).
Integration of multi-omics data and cohort-based analyses
Cholangiocarcinoma (CCA) is a rare, highly aggressive and often fatal tumor of the bile ducts. Cholelithiasis, primary sclerosing cholangitis (PSC), infection with the liver fluke parasites O. viverrini or C. sinensis have been described as risk factors in CCA. Our current research focuses on the molecular profiling of CCA using DNA methylation microarrays and genomic sequencing to define distinct patient subgroups. Recently, we showed that four prognostically relevant patient subgroups of intrahepatic CCA can be distinguished by integrating DNA methylation, genomic alterations, and mutations (Goeppert, Toth et al. Hepatology, 2019). Furthermore, we demonstrated that PSC-associated CCA exhibit a distinct extrahepatic morpho-molecular phenotype, that is independent of the anatomical location of the tumor (Goeppert, Folseraas, Roessler et al. Hepatology, 2020). To pinpoint the development of CCA, we are delineating early precursor lesions and aim at identifying the cell-of-origin of CCA subgroups (Goeppert et al. Gut, 2022).
Development and progression of gallbladder carcinoma
Biliary tract cancer comprises gallbladder cancer (GBC) and CCA. GBC is a rare tumor entity with a prevalence of 2 cases per 100,000 individuals in Europe. However, its incidence is much higher for Native Americans, reaching 23 per 100,000. Due to the low prevalence in Western countries, GBC remains under researched. However, the poor prognosis - reflected by a 2-year survival rate of less than 10% - highlights the need for a deeper understanding for this disease. To identify proteins that involved in the development and progression of GBC, we conducted mass spectrometry-based proteomics of human GBC samples. This approach revealed that CEACAM6 is highly upregulated in GBC compared to normal gallbladder epithelium. Functional and molecular analyses showed that CEACM6 acts as an oncogene via ERK and AKT signaling (Sugiyanto et al. Cell Death Dis, 2024).
Publications
Group members


Eva Eiteneuer (Technician)
Kira Gür (Technician)
Kelis Griesbach (Trainee)
Yueyang Hu (Doctoral student)
Dr. Mario Huerta Casado (Postdoc)
Aslihan Inal (PhD student)
Alina Krumme (PhD student)
Emily Neugum (MD student)
Prof. Stephanie Roessler (Group leader)
Associated Physician Scientists
Dr. Thomas Albrecht
Alphonse Charbel
Carmen Metzger
Dr. Johannes Schreck
Funding
DFG (German Research Foundation)
Deutsche Krebshilfe (German Cancer Aid)
PSC Partners Seeking a Cure
Wilhelm Sander-Stiftung
NCT Heidelberg School of Oncology
Core Facility
Prof. Stephanie Roessler leads the section Laser microdissection (LMD).